Indexing, Slicing and Subsetting DataFrames
Last updated on 2025-10-29 | Edit this page
Overview
Questions
- How can I access specific data within my data set?
- How can Python and Pandas help me to analyse my data?
Objectives
- Describe what 0-based indexing is.
- Manipulate and extract data using column headings and index locations.
- Employ slicing to select sets of data from a DataFrame.
- Employ label and integer-based indexing to select ranges of data in a dataframe.
- Reassign values within subsets of a DataFrame.
- Create a copy of a DataFrame.
- Query / select a subset of data using a set of criteria using the
following operators:
==,!=,>,<,>=,<=. - Locate subsets of data using masks.
- Describe BOOLEAN objects in Python and manipulate data using BOOLEANs.
In the first episode of this lesson, we read a CSV file into a pandas’ DataFrame. We learned how to:
- save a DataFrame to a named object,
- create plots based on the data we loaded into pandas
In this lesson, we will explore ways to access different parts of the data using:
- indexing,
- slicing, and
- subsetting
Loading our data
We will continue to use the samples dataset that we worked with in the last episode. Let’s reopen and read in the data again:
Indexing and Slicing in Python
We often want to work with subsets of a DataFrame object. There are different ways to accomplish this including: using labels (column headings), numeric ranges, or specific x,y index locations.
Selecting data using Labels (Column Headings)
We use square brackets [] to select a subset of a Python
object. For example, we can select all data from a column named
species_id from the samples DataFrame by name.
There are two ways to do this:
PYTHON
# TIP: use the .head() method we saw earlier to make output shorter
# Method 1: select a 'subset' of the data using the column name
samples['species_id']
# Method 2: use the column name as an 'attribute'; gives the same output
samples.species_idWe can pass a list of column names too, as an index to select columns in that order. This is useful when we need to reorganize our data.
NOTE: If a column name is not contained in the DataFrame, an exception (error) will be raised.
PYTHON
# Select the species and plot columns from the DataFrame
samples[['species_id', 'plot_id']]
# What happens when you flip the order?
samples[['plot_id', 'species_id']]
# What happens if you ask for a column that doesn't exist?
samples['speciess']Python tells us what type of error it is in the traceback, at the
bottom it says KeyError: 'speciess' which means that
speciess is not a valid column name (nor a valid key in the
related Python data type dictionary).
Reminder
The Python language and its modules (such as Pandas) define reserved
words that should not be used as identifiers when assigning objects and
variable names. Examples of reserved words in Python include Boolean
values True and False, operators
and, or, and not, among others.
The full list of reserved words for Python version 3 is provided at https://docs.python.org/3/reference/lexical_analysis.html#identifiers.
When naming objects and variables, it’s also important to avoid using
the names of built-in data structures and methods. For example, a
list is a built-in data type. It is possible to use the word
‘list’ as an identifier for a new object, for example
list = ['apples', 'oranges', 'bananas']. However, you would
then be unable to create an empty list using list() or
convert a tuple to a list using list(sometuple).
Extracting Range based Subsets: Slicing
Reminder
Python uses 0-based indexing.
Let’s remind ourselves that Python uses 0-based indexing. This means
that the first element in an object is located at position
0. This is different from other tools like R and Matlab
that index elements within objects starting at 1.
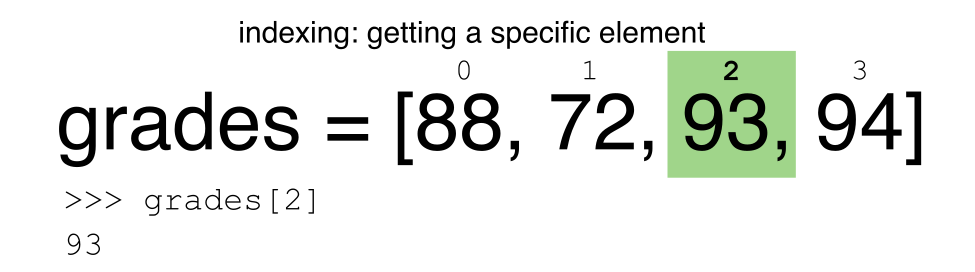
a[0]returns1, as Python starts with element 0 (this may be different from what you have previously experience with other languages e.g. MATLAB and R)a[5]raises anIndexErrorThe error is raised because the list
ahas no element with index 5: it has only five entries, indexed from 0 to 4.-
a[len(a)]also raises anIndexError.len(a)returns5, makinga[len(a)]equivalent toa[5]. To retreive the final element of a list, us the index-1, e.g.OUTPUT
5
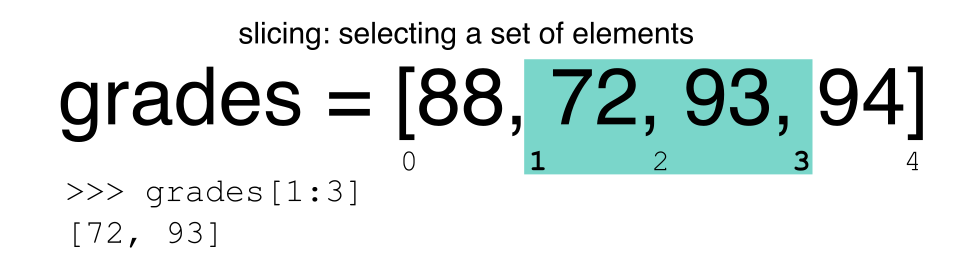
Slicing Subsets of Rows in Python
Slicing using the [] operator selects a set of rows
and/or columns from a DataFrame. To slice out a set of rows, you use the
following syntax: data[start:stop]. When slicing in pandas
the start bound is included in the output. The stop bound is one step
BEYOND the row you want to select. So if you want to select rows 0, 1
and 2 your code would look like this:
The stop bound in Python is different from what you might be used to in languages like Matlab and R.
Slicing Subsets of Rows and Columns in Python
We just learned we can use square brackets to slice our DataFrame and
select columns or rows. And this is useful overall in Python for
selecting a subset of items from a data structure. However, the pandas
library recommends to use .loc[] and .iloc[]
instead. With bot of these we can select specific ranges of our data in
both the row and column directions, using either label or integer-based
indexing.
locis primarily a label based indexing where you can refer to rows and columns by their name. E.g., column ‘month’. Note that integers may be used, but they are interpreted as a label.ilocis primarily an integer based indexing counting from 0. That is, you specify rows and columns using a number. Thus, the first row is row 0, the second column is column 1, etc.
To select a subset of rows and columns from our
DataFrame, we can use the loc method. For example, we can
select the first three rows, and the month, day and year columns, like
this:
which gives the output
OUTPUT
month day year
0 7 16 1977
1 7 16 1977
2 7 16 1977
3 7 16 1977Let’s explore some other ways to index and select subsets of data:
PYTHON
# Select all columns for rows of index values 0 and 10
samples.loc[[0, 10], :]
# What does this do?
samples.loc[0, ['species_id', 'plot_id', 'weight']]
# What happens when you type the code below?
samples.loc[[0, 10, 35549], :]NOTE: Labels must be found in the DataFrame or you
will get a KeyError.
We can also select a specific data value using a row and column
location within the DataFrame and iloc indexing:
In this iloc example,
gives the output
OUTPUT
'F'Remember that Python indexing begins at 0. So, the index location [2, 6] selects the element that is 3 rows down and 7 columns over in the DataFrame.
Challenge - Range
- What happens when you execute:
samples[0:1]samples[0]samples[:4]samples[:-1]
- What happens when you call:
samples.iloc[0:1]samples.iloc[0]samples.iloc[:4, :]samples.iloc[0:4, 1:4]samples.loc[0:4, 1:4]
- How are the last two commands different?
-
-
samples[0:3]returns the first three rows of the DataFrame:
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 0 1 7 16 1977 2 NL M 32.0 NaN 1 2 7 16 1977 3 NL M 33.0 NaN 2 3 7 16 1977 2 DM F 37.0 NaN-
samples[0]results in a ‘KeyError’, since direct indexing of a row is redundant withiloc. -
samples[0:1]can be used to obtain only the first row. -
samples[:5]slices from the first row to the fifth:
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 0 1 7 16 1977 2 NL M 32.0 NaN 1 2 7 16 1977 3 NL M 33.0 NaN 2 3 7 16 1977 2 DM F 37.0 NaN 3 4 7 16 1977 7 DM M 36.0 NaN 4 5 7 16 1977 3 DM M 35.0 NaN-
samples[:-1]provides everything except the final row of the DataFrame. You can use negative index numbers to count backwards from the last entry.
-
-
samples.iloc[0:1]returns the first row -
samples.iloc[0]returns the first row as a named list -
samples.iloc[:4, :]returns all columns of the first four rows -
samples.iloc[0:4, 1:4]selects specified columns of the first four rows -
samples.loc[0:4, 1:4]results in a ‘TypeError’ - see below.
-
- While
ilocuses integers as indices and slices accordingly,locworks with labels. It is like accessing values from a dictionary, asking for the key names. Column names 1:4 do not exist, so the call tolocabove results in an error. Check also the difference betweensamples.loc[0:4]andsamples.iloc[0:4].
Subsetting Data using Criteria
We can also select a subset of our data using criteria. For example, we can select all rows that have a year value of 2002:
Which produces the following output:
PYTHON
record_id month day year plot_id species_id sex hindfoot_length \
9789 9790 1 19 1985 16 RM F 16.0
9790 9791 1 19 1985 17 OT F 20.0
9791 9792 1 19 1985 6 DO M 35.0
9792 9793 1 19 1985 12 DO F 35.0
9793 9794 1 19 1985 24 RM M 16.0
... ... ... ... ... ... ... ... ...
11222 11223 12 8 1985 4 DM M 36.0
11223 11224 12 8 1985 11 DM M 37.0
11224 11225 12 8 1985 7 PE M 20.0
11225 11226 12 8 1985 1 DM M 38.0
11226 11227 12 8 1985 15 NaN NaN NaN
weight genus species taxa plot_type
9789 4.0 Reithrodontomys megalotis Rodent Rodent Exclosure
9790 16.0 Onychomys torridus Rodent Control
9791 48.0 Dipodomys ordii Rodent Short-term Krat Exclosure
9792 40.0 Dipodomys ordii Rodent Control
9793 4.0 Reithrodontomys megalotis Rodent Rodent Exclosure
... ... ... ... ... ...
11222 40.0 Dipodomys merriami Rodent Control
11223 49.0 Dipodomys merriami Rodent Control
11224 18.0 Peromyscus eremicus Rodent Rodent Exclosure
11225 47.0 Dipodomys merriami Rodent Spectab exclosure
11226 NaN NaN NaN NaN Long-term Krat Exclosure
[1438 rows x 13 columns]Or we can select all rows that do not contain the year 2002:
We can define sets of criteria too:
Python Syntax Cheat Sheet
We can use the syntax below when querying data by criteria from a DataFrame. Experiment with selecting various subsets of the “samples” data.
- Equals:
== - Not equals:
!= - Greater than:
> - Less than:
< - Greater than or equal to:
>= - Less than or equal to:
<=
Challenge - Queries
Select a subset of rows in the
samplesDataFrame that contain data from the year 1999 and that contain weight values less than or equal to 8. How many rows did you end up with? What did your neighbor get?-
You can use the
isincommand in Python to query a DataFrame based upon a list of values as follows:Use the
isinfunction to find all plots that contain the particular speciesNLandDMin the “samples” DataFrame. How many records contain these values? Experiment with other queries. Create a query that finds all rows with a weight value greater than or equal to 0.
The
~symbol in Python can be used to return the OPPOSITE of the selection that you specify. It is equivalent to is not in. Write a query that selects all rows with sex NOT equal to ‘M’ or ‘F’ in the “samples” data.
-
If you are only interested in how many rows meet the criteria, the sum of
Truevalues could be used instead:OUTPUT
63 -
For example, using
NLandDM:OUTPUT
array([ 2, 3, 7, 1, 8, 6, 4, 14, 15, 13, 9, 11, 10, 17, 20, 23, 21, 18, 22, 19, 12, 5, 16, 24])OUTPUT
(24,) samples.loc[samples["weight"] >= 0, :]
Using masks to identify a specific condition
A mask can be useful to locate where a particular
subset of values exist or don’t exist - for example, NaN, or “Not a
Number” values. To understand masks, we also need to understand
BOOLEAN objects in Python.
Boolean values include True or False. For
example,
When we ask Python whether x is greater than 5, it
returns False. This is Python’s way to say “No”. Indeed,
the value of x is 5, and 5 is not greater than 5.
To create a boolean mask:
- Set the True / False criteria
(e.g.
values > 5 = True) - Python will then assess each value in the object to determine whether the value meets the criteria (True) or not (False).
- Python creates an output object that is the same shape as the
original object, but with a
TrueorFalsevalue for each index location.
Let’s try this out. Let’s identify all locations in the survey data
that have null (missing or NaN) data values. We can use the
isna method to do this. The isna method will
compare each cell with a null value. If an element has a null value, it
will be assigned a value of True in the output object.
A snippet of the output is below:
PYTHON
record_id month day year plot_id species_id sex \
0 False False False False False False False
1 False False False False False False False
2 False False False False False False False
3 False False False False False False False
4 False False False False False False False
... ... ... ... ... ... ... ...
16873 False False False False False False False
16874 False False False False False False False
16875 False False False False False False False
16876 False False False False False False False
16877 False False False False False False False
hindfoot_length weight genus species taxa plot_type
0 False True False False False False
1 False True False False False False
2 False True False False False False
3 False True False False False False
4 False True False False False False
... ... ... ... ... ... ...
16873 False False False False False False
16874 False False False False False False
16875 False False False False False False
16876 False False False False False False
16877 False False False False False False
[16878 rows x 13 columns]To select the rows where there are null values, we can use the mask as an index to subset our data as follows:
PYTHON
# To select just the rows with NaN values, we can use the 'any()' method
samples.loc[samples.isna().any(axis=1), :]We can run isna on a particular column too. What does
the code below do?
PYTHON
# What does this do?
empty_weights = samples.loc[samples["weight"].isna(), "weight"]
print(empty_weights)Let’s take a minute to look at the statement above. We are using the
Boolean object samples["weight"].isna() as an index to
samples. We are asking Python to select rows that have a
NaN value of weight.
Challenge - Putting it all together
Create a new DataFrame that only contains observations with sex values that are not female or male. Print the number of rows in this new DataFrame. Verify the result by comparing the number of rows in the new DataFrame with the number of rows in the samples DataFrame where sex is null.
- In Python, portions of data can be accessed using indices, slices, column headings, and condition-based subsetting.
- Python uses 0-based indexing, in which the first element in a list, tuple or any other data structure has an index of 0.
- Pandas enables common data exploration steps such as data indexing, slicing and conditional subsetting.
