Content from Getting Started
Last updated on 2025-10-27 | Edit this page
Overview
Questions
- How can I identify and use key features of JupyterLab to create and manage a Python notebook?
- How do I run Python code in JupyterLab, and how can I see and interpret the results?
Objectives
- Launch JupyterLab and create a new Jupyter Notebook.
- Navigate the JupyterLab interface, including file browsing, cell creation, and cell execution, with confidence.
- Write and execute Python code in a Jupyter Notebook cell, observing the output and modifying code as needed.
- Save a Jupyter Notebook as an .ipynb file and verify the file’s location in the directory within the session.
This episode is a modified copy of the Getting Started episode of the Python Intro for Libraries lesson.
Why Python?
Python is a popular programming language for tasks such as data collection, cleaning, and analysis. Python can help you to create reproducible workflows to accomplish repetitive tasks more efficiently.
What is Python?
Python is a general purpose programming language that supports rapid development of data analytics applications. The word “Python” is used to refer to both, the programming language and the tool that executes the scripts written in Python language.
Its main advantages are:
- Free
- Open-source
- Available on all major platforms (macOS, Linux, Windows)
- Supported by Python Software Foundation
- Supports multiple programming paradigms
- Has large community
- Rich ecosystem of third-party packages
So, why do you need Python for data analysis?
Easy to learn: Python is easier to learn than other programming languages. This is important because lower barriers mean it is easier for new members of the community to get up to speed.
-
Reproducibility: Reproducibility is the ability to obtain the same results using the same dataset(s) and analysis.
Data analysis written as a Python script can be reproduced on any platform. Moreover, if you collect more or correct existing data, you can quickly re-run your analysis!
An increasing number of journals and funding agencies expect analyses to be reproducible, so knowing Python will give you an edge with these requirements.
-
Versatility: Python is a versatile language that integrates with many existing applications to enable something completely amazing. For example, one can use Python to generate manuscripts, so that if you need to update your data, analysis procedure, or change something else, you can quickly regenerate all the figures and your manuscript will be updated automatically.
Python can read text files, connect to databases, and many other data formats, on your computer or on the web.
Interdisciplinary and extensible: Python provides a framework that allows anyone to combine approaches from different research (but not only) disciplines to best suit your analysis needs.
Python has a large and welcoming community: Thousands of people use Python daily. Many of them are willing to help you through mailing lists and websites, such as [Stack Overflow][stack-overflow] and [Anaconda community portal][anaconda-community].
Free and Open-Source Software (FOSS)… and Cross-Platform: We know we have already said that but it is worth repeating.
Research Project: Best Practices
It is a good idea to keep a set of related data, analyses, and text in a single folder. All scripts and text files within this folder can then use relative paths to the data files. Working this way makes it a lot easier to move around your project and share it with others.
Organizing your working directory
Using a consistent folder structure across your projects will help you keep things organized, and will also make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you may wish to create separate directories for your scripts, data, and documents.
data/: Use this folder to store your raw data. For the sake of transparency and provenance, you should always keep a copy of your raw data. If you need to cleanup data, do it programmatically (i.e. with scripts) and make sure to separate cleaned up data from the raw data. For example, you can store raw data in files./data/raw/and clean data in./data/clean/.documents/: Use this folder to store outlines, drafts, and other text.code/: Use this folder to store your (Python) scripts for data cleaning, analysis, and plotting that you use in this particular project.
You may need to create additional directories depending on your
project needs, but these should form the backbone of your project’s
directory. For this workshop, we will need a data/ folder
to store our raw data, and we will later create a
data_output/ folder when we learn how to export data as CSV
files.
Use JupyterLab to edit and run Python code.
For having a common platform to run our Python code, we’ll use the JupyterHub instance provided by LSIT. If you want to install Python on your machine, the setup instructions for details on how to install Python, JupyterLab, and the necessary packages using Pixi.
Getting started with JupyterLab
To run Python, we are going to use Jupyter Notebooks via JupyterLab. Jupyter notebooks are common tools for data science and visualization, and serve as a convenient environment for running Python code interactively where we can view and share the results of our Python code.
Alternatives to Juypter
There are other ways of editing, managing, and running Python code. Software developers often use an integrated development environment (IDE) like PyCharm, Spyder or Visual Studio Code (VS Code), to create and edit Python scripts. Others use text editors like Vim or Emacs to hand-code Python. After editing and saving Python scripts you can execute those programs within an IDE or directly on the command line.
Jupyter notebooks let us execute and view the results of our Python code immediately within the notebook. JupyterLab has several other handy features:
- You can easily type, edit, and copy and paste blocks of code.
- It allows you to annotate your code with links, different sized text, bullets, etc. to make it more accessible to you and your collaborators.
- It allows you to display figures next to the code to better explore your data and visualize the results of your analysis.
- Each notebook contains one or more cells that contain code, text, or images.
The JupyterLab Interface
Launching JupyterLab opens a new tab or window in your preferred web
browser. While JupyterLab enables you to run code from your browser, it
does not require you to be online. If you take a look at the URL in your
browser address bar, you should see that the environment is located at
your localhost, meaning it is running from your computer:
http://localhost:8888/lab.
When you first open JupyterLab you will see two main panels. In the
left sidebar is your file browser. You should see a folder in the file
browser named data that contains all of our data.
Creating a Juypter Notebook
To the right you will see a Launcher tab. Here we have
options to launch a Python 3 notebook, a Terminal (where we can use
shell commands), text files, and other items. For now, we want to launch
a new Python 3 notebook, so click once on the
Python 3 (ipykernel) button underneath the Notebook header.
You can also create a new notebook by selecting New ->
Notebook from the File menu in the Menu Bar.
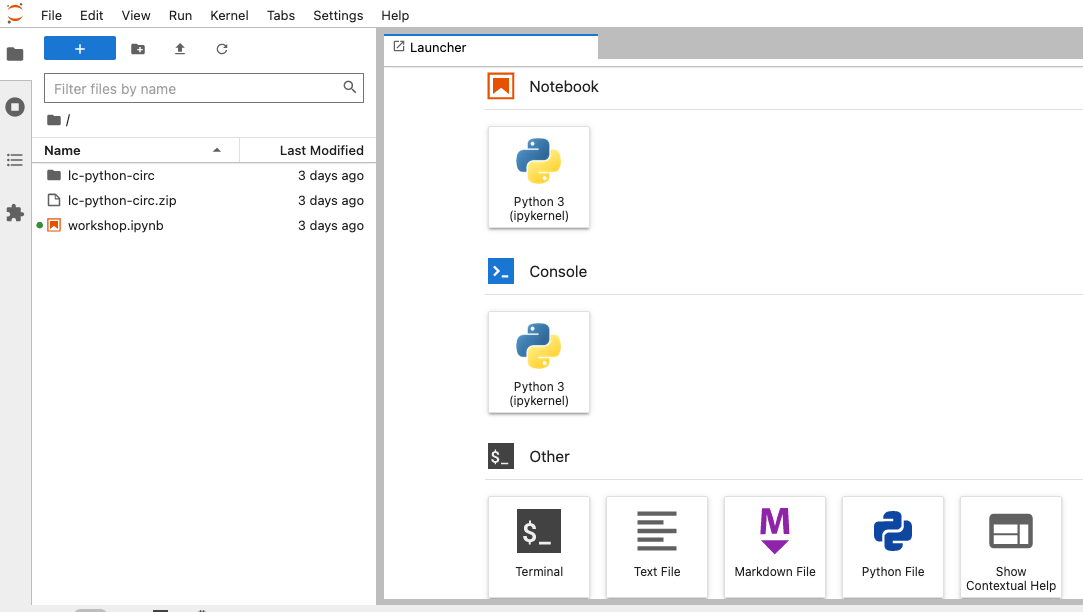
When you start a new Notebook you should see a new tab labeled
Untitled.ipynb. You will also see this file listed in the
file browser to the left. Right-click on the Untitled.ipynb
file in the file browser and choose Rename from the
dropdown options. Let’s call the notebook file,
workshop.ipynb.
JupyterLab? What about Jupyter notebooks? Python notebooks? IPython?
JupyterLab is the next stage in the evolution of the Jupyter Notebook. If you have prior experience working with Jupyter notebooks, then you will have a good idea of how to work with JupyterLab. Jupyter was created as a spinoff of IPython in 2014, and includes interactive computing support for languages other than just Python, including R and Julia. While you’ll still see some references to Python and IPython notebooks, IPython notebooks are officially deprecated in favor of Jupyter notebooks.
We will share more features of the JupyterLab environment as we advance through the lesson, but for now let’s turn to how to run Python code.
Running Python code
Jupyter allows you to add code and formatted text in different types of blocks called cells. By default, each new cell in a Jupyter Notebook will be a “code cell” that allows you to input and run Python code. Let’s start by having Python do some arithmetic for us.
In the first cell type 7 * 3, and then press the
Shift+Return keys together to execute the contents
of the cell. (You can also run a cell by making sure your cursor is in
the cell and choosing Run > Run Selected Cells or
selecting the “Play” icon (the sideways triangle) at the top of the
noteboook.)
You should see the output appear immediately below the cell, and Jupyter will also create a new code cell for you.
If you move your cursor back to the first cell, just after the
7 * 3 code, and hit the Return key (without
shift), you should see a new line in the cell where you can add more
Python code. Let’s add another calculation to the same cell:
While Python runs both calculations Juypter will only display the output from the last line of code in a specific cell, unless you tell it to do otherwise.
Editing the notebook
You can use the icons at the top of your notebook to edit the cells in your Notebook:
- The
+icon adds a new cell below the selected cell. - The scissors icon will delete the current cell.
You can move cells around in your notebook by hovering over the left-hand margin of a cell until your cursor changes into a four-pointed arrow, and then dragging and dropping the cell where you want it.
Markdown
You can add text to a Juypter notebook by selecting a cell, and
changing the dropdown above the notebook from Code to
Markdown. Markdown is a lightweight language for formatting
text. This feature allows you to annotate your code, add headers, and
write documentation to help explain the code. While we won’t cover
Markdown in this lesson, there are many helpful online guides out there:
- Markdown
for Jupyter Cheatsheet (IBM) - Markdown Guide (Matt Cone)

You can also use “hotkeys”” to change Jupyter cells from Code to Markdown and back:
- Click on the code cell that you want to convert to a Markdown cell.
- Press the Esc key to enter command mode.
- Press the M key to convert the cell to Markdown.
- Press the y key to convert the cell back to Code.
- You can launch JupyterLab from the command line or from Anaconda Navigator.
- You can use a JupyterLab notebook to edit and run Python.
- Notebooks can include both code and markdown (text) cells.
Content from Data visualization with Pandas and Matplotlib
Last updated on 2025-10-28 | Edit this page
Overview
Questions
- How do you start exploring and visualizing data using Python?
- How can you make and customize plots?
Objectives
- Explain the difference between installing a library and importing it.
- Load data from a CSV file into a pandas DataFrame and inspect its contents and structure.
- Generate plots, such as scatter plots and box plots, directly from a pandas DataFrame.
- Construct a Matplotlib figure containing multiple subplots.
- Customize plot aesthetics like titles, axis labels, colors, and layout by passing arguments to plotting functions.
- Export a completed figure to a file.
Loading libraries
A great feature in Python is the ability to import libraries to extend its capabilities. For now, we’ll focus on two of the most widely used libraries for data analysis: pandas and Matplotlib. We’ll be using pandas for data wrangling and manipulation, and Matplotlib for (you guessed it) making plots.
To be able to use these libraries in our code, we have to install and import them. Installation is needed as pandas and Matplotlib are third-party libraries that aren’t built into Python. You should have gone through the installation process during the setup for the workshop (if not, visit the setup page), so we’ll jump straight to showing you how to import libraries.
To import a library, we use the syntax
import libraryName. If we want to give the library a
nickname to shorten the command each time we call it, we can add
as nickNameHere. Here is how you’d import pandas and
Matplotlib using the common nicknames pd and
plt, respectively.
If you got an error similar to
ModuleNotFoundError: No module named '___', it means you
haven’t installed the library. Check the setup
page to install it and double check you typed it correctly.
If you’re asking yourself why we used matplotlib.pyplot
instead of just matplotlib, good question! We are not
importing the entire Matplotlib library, we are only importing the
fraction of it we need for plotting, called pyplot. We will return to
this topic later to explain more about matplotlib.pyplot
and why that is the part that we need to import.
For the purposes of this workshop, you only need to install a library once on your computer. However, you must import it in every notebook or script where you plan to use it, since Python doesn’t automatically load installed libraries.
In your future work with Python, this may not always be the case. You might want to keep different projects separate by using a different Python environment for each one. In that case, you’ll need to install the same library in each environment. We’ll talk more about environments later in the workshop.
Loading and exploring data
For this lesson, we will be using the Portal Teaching data, a subset of the data from Ernst et al. Long-term monitoring and experimental manipulation of a Chihuahuan Desert ecosystem near Portal, Arizona, USA.
We will be using files from the Portal
Project Teaching Database. This section will use the
surveys_complete_77_89.csv file that can be downloaded
here: https://datacarpentry.github.io/R-ecology-lesson/data/cleaned/surveys_complete_77_89.csv
We are studying the weight, hindfoot length, and sex of animals
caught in sites of our study area. The data is stored as a
.csv file: each row holds information for a single animal,
and the columns represent:
| Column | Description |
|---|---|
| record_id | Unique id for the observation |
| month | month of observation |
| day | day of observation |
| year | year of observation |
| plot_id | ID of a particular site |
| species_id | 2-letter code |
| sex | sex of animal (“M”, “F”) |
| hindfoot_length | length of the hindfoot in mm |
| weight | weight of the animal in grams |
| genus | the genus of the species |
| species | the latin species name |
| taxa | general taxonomic category |
| plot_type | type of experimental manipulation conducted |
We’ll load the data in the csv into Python and name this new object
samples. For this, we can use the pandas
library and its .read_csv() function, like shown here:
Here we have created a new object that we can reference later in our
code. All objects in Python have a type, which determine
what is possible to do with them. To know the type of an object, we can
use the type() function.
OUTPUT
pandas.core.frame.DataFrameNotice how we didn’t use the = operator in this case.
This means we didn’t create a new object, we just asked Python to show
us the output of the function. When we created samples with
the = operator it didn’t print an output, but it stored the
object in Python which we can later use.
From the output we can read that samples is an object of
pandas DataFrame type. We’ll explore in depth what a DataFrame is in the
next episode. For now, we only need to keep in mind that our data is now
contained in a DataFrame. And this is important as the methods we’ll
cover now -.head(), .info(), and
.plot()- are only available to DataFrame objects.
Now that our data is in Python, we can start working with it. A good
place to start is taking a look at the data. With the
.head() method, we can see the first five rows of the data
set.
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length \
0 1 7 16 1977 2 NL M 32.0
1 2 7 16 1977 3 NL M 33.0
2 3 7 16 1977 2 DM F 37.0
3 4 7 16 1977 7 DM M 36.0
4 5 7 16 1977 3 DM M 35.0
weight genus species taxa plot_type
0 NaN Neotoma albigula Rodent Control
1 NaN Neotoma albigula Rodent Long-term Krat Exclosure
2 NaN Dipodomys merriami Rodent Control
3 NaN Dipodomys merriami Rodent Rodent Exclosure
4 NaN Dipodomys merriami Rodent Long-term Krat Exclosure The .tail() method will give us instead the last five
rows of the data. If we want to override this default and instead
display the last two rows, we can use the n argument. An
argument is an input that a function or method takes to
modify how it operates, and you set arguments using the =
sign.
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length \
16876 16877 12 5 1989 11 DM M 37.0
16877 16878 12 5 1989 8 DM F 37.0
weight genus species taxa plot_type
16876 50.0 Dipodomys merriami Rodent Control
16877 42.0 Dipodomys merriami Rodent ControlAnother useful method to get a glimpse of the data is
.info(). This tells us how many rows the data set has (# of
entries), the number of columns, and for each of the columns it says its
name, the number of non-null (or non-empty) rows it contains, and its
data type.
OUTPUT
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 16878 entries, 0 to 16877
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 record_id 16878 non-null int64
1 month 16878 non-null int64
2 day 16878 non-null int64
3 year 16878 non-null int64
4 plot_id 16878 non-null int64
5 species_id 16521 non-null object
6 sex 15578 non-null object
7 hindfoot_length 14145 non-null float64
8 weight 15186 non-null float64
9 genus 16521 non-null object
10 species 16521 non-null object
11 taxa 16521 non-null object
12 plot_type 16878 non-null object
dtypes: float64(2), int64(5), object(6)
memory usage: 1.7+ MBFunctions and methods in Python
As we saw, functions allow us to perform a given task. They take
inputs, perform the task on those inputs, and return an output. For
example, with the pd.read_csv() function, we gave the file
path as input, and it returned the DataFrame as output.
Functions can be built-in, which means they come natively with
Python, like type() or print(). Or they can
come from an imported library, like pd.read_csv() comes
from Pandas.
A method is similar to a function. The only difference is a method is
associated with a given object type. That’s why we use the syntax
object_name.method_name().
This is the case for .head(), .tail(), and
.info(): they are methods that the pandas
DataFrame object carries around with it.
Basic plotting with Pandas
pandas makes it easy to start plotting your data with the
.plot() method. By passing arguments to this method, we can
tell Pandas how we want the plot to look.
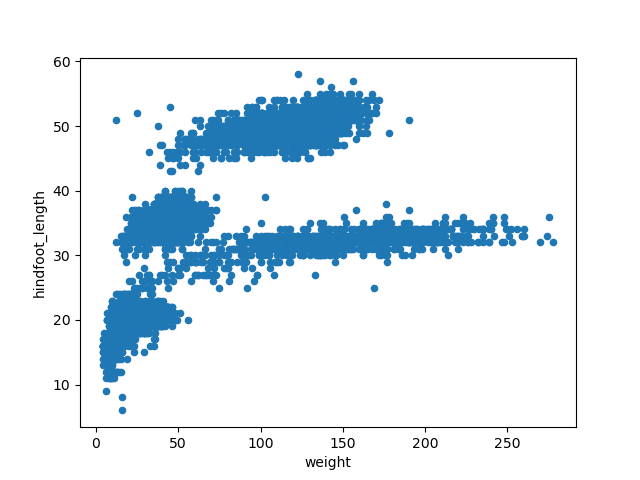
With the following code, we will make a scatter plot (argument
kind = 'scatter') to analyze the relationship between the
weight (which will plot in the x axis, argument
x = 'weight') and the hindfoot length (in the y axis,
argument y = 'hindfoot_length') of the animals sampled at
the study site.
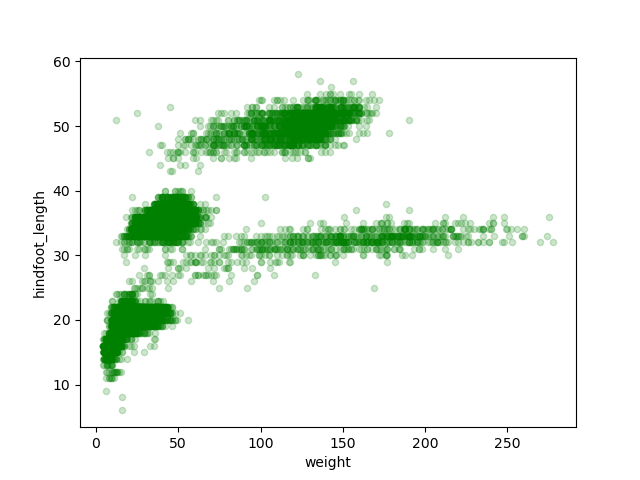
When coding, you’ll often find the case where you can get to the same
result using different code. In this case, the creators of pandas make
it possible to make the previous plot with the.plot.scatter
method, without having to specify the “kind” argument.
This scatter plot shows there seems to be a positive relation between
weight and hindfoot length, where heavier animals tend to have bigger
hindfeet. But you may have noticed that parts of our scatter plot have
many overlapping points, making it difficult to see all the data. We can
adjust the transparency of the points using the alpha
argument, which takes a value between 0 and 1.
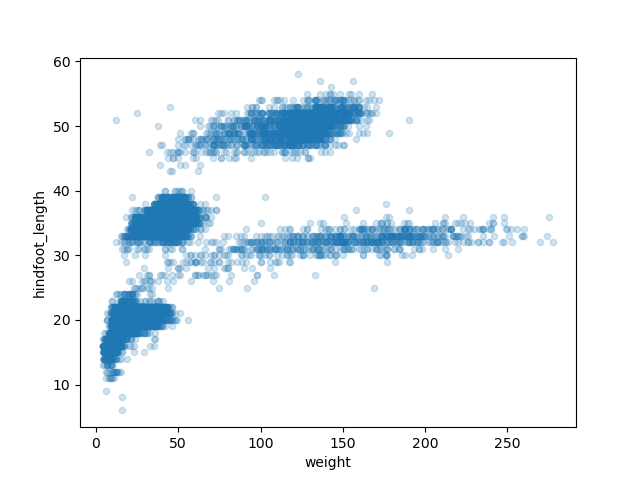
With transparency added to the points, we can more clearly observe a clustering of data points into several more densely populated regions of the scatter plot.
There are multiple ways we can learn what other arguments we have available to modify the looks of our plot. One of those ways is reading the documentation of the library we are using. In the next episode, we’ll cover other ways to get help in Python.
Challenge - Changing the color of points
Check the documentation of the pandas method
.plot.scatter() to learn what argument you can use to
change the color of the points in the scatter plot to green.
Here is the link to the documentation: https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.plot.scatter.html
Similarly, we could make a box plot to explore
the distribution of the hindfoot length across all samples. In this
case, pandas wants us to use different arguments. We’ll use the
column argument to specify what is the column we want to
analyze.
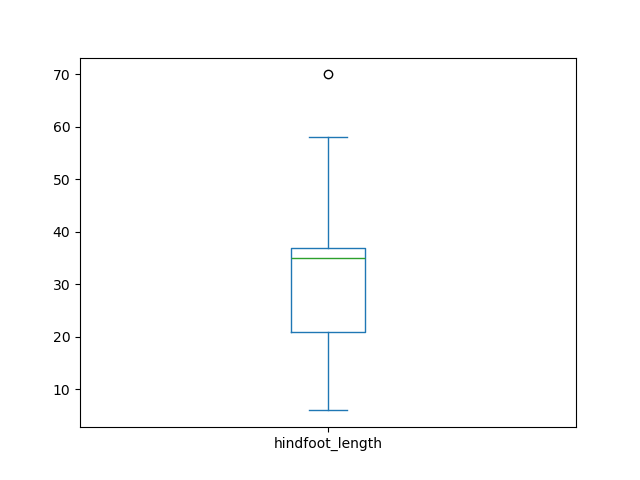
The box plot shows the median hindfoot length is around 32mm (represented by the line inside the box) and most values lie between 20 and 35 mm (which are the borders of the box, representing the 1st and 3rd quartile of the data, respectively).
We could further expand this analysis, and see the distribution of
this variable across different plot types. We can add a by
argument, saying by which variable we want do disaggregate the box
plot.

As shown in the previous image, the x-axis labels overlap with each other, which makes them unreadable. Furthermore, we’d like to start customizing the title and the axis labels. Or maybe make multiple subplots in the same figure. At this point we realize a more fine-grained control over our graph is needed, and here is where Matplotlib appears in the picture.
Advanced plots with Matplotlib
Matplotlib is a Python library that is widely used throughout the scientific Python community to create high-quality and publication-ready graphics. It supports a wide range of raster and vector graphics formats including PNG, PostScript, EPS, PDF and SVG.
Moreover, Matplotlib is the actual engine behind the plotting
capabilities of Pandas, and other plotting libraries like seaborn and plotnine. For example, when we call the
.plot() methods on pandas data objects, Matplotlib is
actually being used “backstage”.
Our first step in the process is creating our figure and our axes (or
plots), using the plt.subplots() function.
The fig object we are creating is the entire plot area,
which can contain one or multiple axes. In this case, the function
default is to create only one set of axes, which will be referenced as
the axis object. For now, this results in an empty plot
like a blank canvas for us to start plotting data on to.
We’ll add the previous box plot we made to this axis, by
using the ax argument inside the .plot()
function. This gives us a plot just as we had it before, but now onto an
axis object that we can start customizing.
PYTHON
fig, axis = plt.subplots()
samples.plot(column = 'hindfoot_length', by = 'plot_type',
kind = 'box', ax = axis)
To start, we can rotate the x-axis labels 90 degrees to make them
readable. For this, we use the .tick_params() method on the
axis object.
PYTHON
fig, axis = plt.subplots()
samples.plot(column = 'hindfoot_length', by = 'plot_type',
kind = 'box', ax = axis)
axis.tick_params(axis = 'x', rotation = 90)
Axis objects have a lot of methods like .tick_params(),
which can be used to adjust the layout and styling of the plot. For
example, we can modify the title (the column name is the default) and
add the x- and y-axis labels with the .set_title(),
.set_xlabel(), and .set_ylabel() methods. By
now, you may have begun copying and pasting the parts of the code that
don’t change to save yourself some typing time. Some lines might only
include subtle changes, so take care not to miss anything small and
important when reusing lines written previously.
PYTHON
fig, axis = plt.subplots()
samples.plot(column = 'hindfoot_length', by = 'plot_type',
kind = 'box', ax = axis)
axis.tick_params(axis='x', rotation = 90)
axis.set_title('Distribution of hindfoot lenght across plot types')
axis.set_xlabel('Plot Types')
axis.set_ylabel('Hindfoot Length (mm)')
Making multiple subplots
If we want more than one plot in the same figure, we could specify
the number of rows (nrows argument) and the number of
columns (ncols) when calling the plt.subplots
function. For example, let’s say we want two plots (or axes), organized
in two columns and one row. This will be useful in a minute, when we
arrange our scatter plot and box plot in a single figure.
PYTHON
fig, axes = plt.subplots(nrows = 1, ncols = 2) # note the variable name is 'axes' here rather than 'axis' used above
The axes object contains two objects, which correspond
to the two sets of axes. We can access each of the objects inside
axes using axes[0] and
axes[1].
We’ll get more comfortable with this Python syntax during the workshop, but the most important thing to know now is that Python indexes elements starting with 0. This means the first element in a sequence has index 0, the second element has index 1, the third element is 2, and so forth.
Here is the code to have two subplots, one with the scatter plot (on
axes[0]), and one with the box plot (on
axes[1]):
PYTHON
fig, axes = plt.subplots(nrows = 1, ncols = 2)
samples.plot(x = 'weight', y = 'hindfoot_length', kind='scatter',
alpha = 0.2, ax = axes[0])
samples.plot(column = 'hindfoot_length', by = 'plot_type',
kind = 'box', ax = axes[1])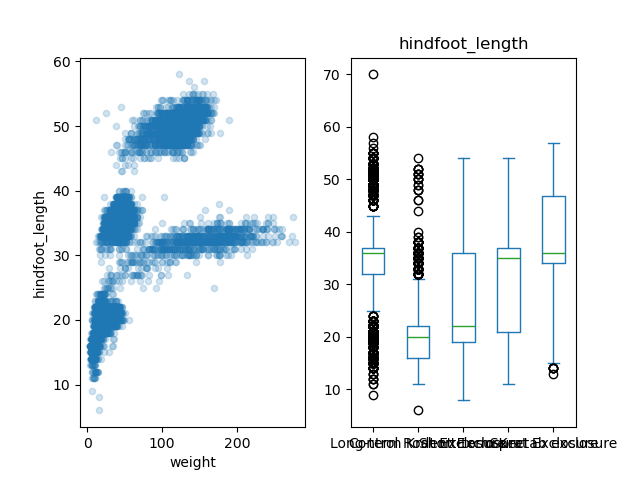
As shown before, Matplotlib allows us to customize every aspect of
our figure. So we’ll make it more professional by adding titles and axis
labels. Notice at the end the introduction of the
.suptitle() method, which allows us to add a super title to
the entire figure. We also use the .tight_layout() method,
to automatically adjust the padding between and around subplots. (Feel
free to try the code below with and without the
fig.tight_layout() line to better understand the difference
it makes.)
PYTHON
fig, axes = plt.subplots(nrows = 1, ncols = 2)
samples.plot(x = 'weight', y = 'hindfoot_length', kind='scatter', alpha = 0.2, ax = axes[0])
axes[0].set_title('Weight vs. Hindfoot Length')
axes[0].set_xlabel('Weight (g)')
axes[0].set_ylabel('Hindfoot Length (mm)')
samples.plot(column = 'hindfoot_length', by = 'plot_type',
kind = 'box', ax = axes[1])
axes[1].tick_params(axis = 'x', rotation = 90)
axes[1].set_title('Hindfoot Length by Plot Type')
axes[1].set_xlabel('Plot Types')
axes[1].set_ylabel('Hindfoot Length (mm)')
fig.suptitle('Analysis of Hindfoot Length variable', fontsize=16)
fig.tight_layout()
You now have the basic tools to create and customize plots using pandas and Matplotlib! Let’s put it into practice.
Challenge - Changing figure size
Our plot probably needs more vertical space, so let’s change the figure size. Take a look at Matplotlib’s figure documentation and answer:
- What is the argument we need to make this adjustment?
- What is the default figure size if we don’t change the argument?
- In which units is the size specified (inches, pixels, centimeters)?
- Use the argument in the
plt.subplots()function to make the previous figure 7x7 inches in size.
The argument we’re looking for is figsize. Figure
dimension is specified as (width, height) in inches, and the default
figure size is 6.4 inches wide and 4.8 inches tall. We can add the
figsize = (7,7) argument like this:
And keep the rest of our plot the same.
Challenge - Add a third axis
Let’s explore the data for the hindfoot length variable more deeply.
Add a third axis to the plot we’ve been working on, for a histogram of
the hindfoot length. For this you can add the argument
kind = hist to an additional .plot() method
call.
After that, there’s a few things you can do to make this graph look nicer. Try making each of these one at a time to see how it changes.
- Increase the size of the figure to make it 10in wide and 7in long, just like in the previous challenge.
- Make the histogram orientation horizontal and hide the legend. For
this, add the
orientation='horizontal'andlegend = Falsearguments to your.plot()method. - As the y-axis of each subplot is the same, we could use a shared y-axis for all three. Explore the Matplotlib documentation some more to find out which parameter you can use to achieve this.
- Add a nice title to your third subplot.
Here is the final code to achieve all of the previous goals. Notice what has changed from our previous code.
PYTHON
fig, axes = plt.subplots(nrows = 1, ncols = 3, figsize = (10,7), sharey = True)
samples.plot(x = 'weight', y = 'hindfoot_length', kind='scatter',
alpha = 0.2, ax = axes[0])
axes[0].set_title('Weight vs. Hindfoot Length')
axes[0].set_xlabel('Weight (g)')
axes[0].set_ylabel('Hindfoot Length (mm)')
samples.plot(column = 'hindfoot_length', by = 'plot_type',
kind = 'box', ax = axes[1])
axes[1].tick_params(axis = 'x', rotation = 90)
axes[1].set_title('Hindfoot Length by Plot Type')
axes[1].set_xlabel('Plot Types')
axes[1].set_ylabel('Hindfoot Length (mm)2')
samples.plot(column = 'hindfoot_length', orientation='horizontal',
legend = False, kind = 'hist', ax = axes[2])
axes[2].set_title('Hindfoot Length Histogram')
fig.suptitle('Analysis of Hindfoot Length variable', fontsize=16)
fig.tight_layout()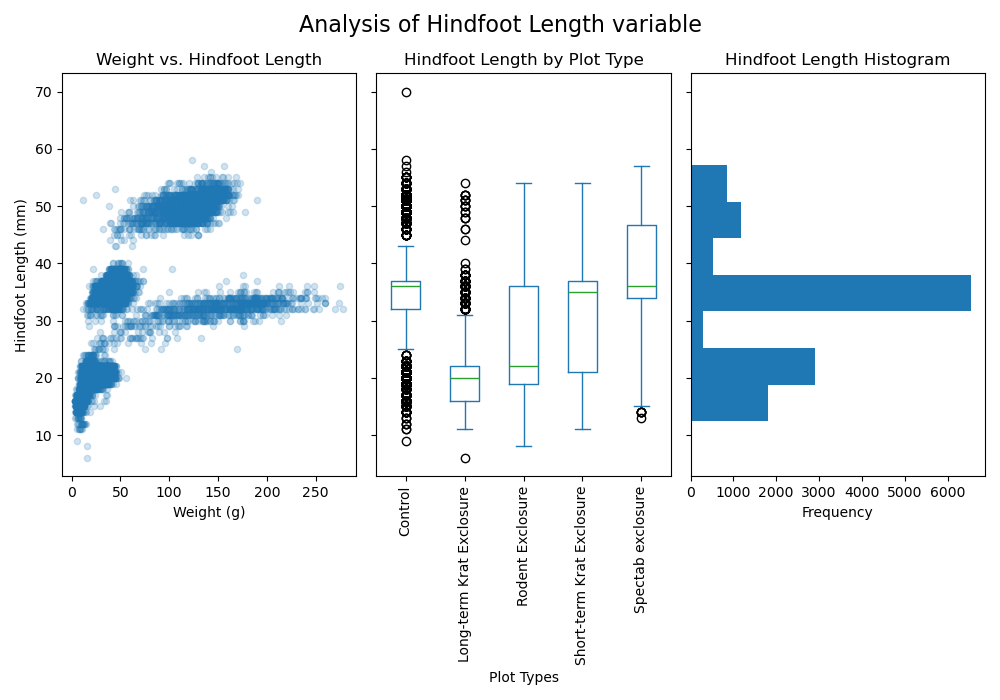
Exporting plots
Until now our plots only live inside Python. Therefore, once we’re
satisfied with the resulting plot, we need to save it with the
.savefig() method on our figure object. The only required
argument is the file path in your computer where you want to save it.
Matplotlib recognizes the extension used in the filename and supports
(on most computers) png, pdf, ps, eps and svg formats.
Challenge - Make and save your own plot
Put all this knowledge to practice and come up with your own plot of
the Portal Teaching data set. Use a different combination of variables,
or a different kind of plot. Here is the pandas
pd.plot() function documentation, where you can see
what other values you can use in the kind argument.
Save your plot to the “images” folder with a .pdf extension.
Other plotting libraries
Learning to plot with pandas and Matplotlib can be overwhelming by itself. However, we want to invite you to explore other plotting libraries available in Python, as some types of plots will be easier to make with them. This will also take time and practice, as every new library come with its own functions, methods, and arguments. But with time and practice, you’ll get used to researching and learning about these new libraries and how they work.
This is where the true power of open source software lies. There is the big community of contributors who all the time are creating new libraries or improving the existing ones.
Some of the libraries you might find useful are:
-
Plotnine: Inspired by R’s ggplot2, it implements the “grammar of graphics” approach. If you come from Rworld and the tidyverse, you’ll definitely want to check it out.
On a plotnine figure, a Matplotlib figure is returned when you use the
.draw()method, so you can always start with plotnine and then customize using Matplotlib. -
Seaborn: Focusing on statistical graphics, there’s a variety of plot types that will be simpler to make (require fewer lines of code) in seaborn. To name a few: statistical representations of averages or of simple linear regression models; plots of categorical data; multivariate plots, etc. Refer to the introduction to seaborn to learn more.
For example, the following scatter plot which adds “sex” as a third variable would require more lines of code if done with pandas + Matplotlib alone.
PYTHON
import seaborn as sns sns.scatterplot(data = samples, x='weight', y='hindfoot_length', hue='sex', alpha = 0.2)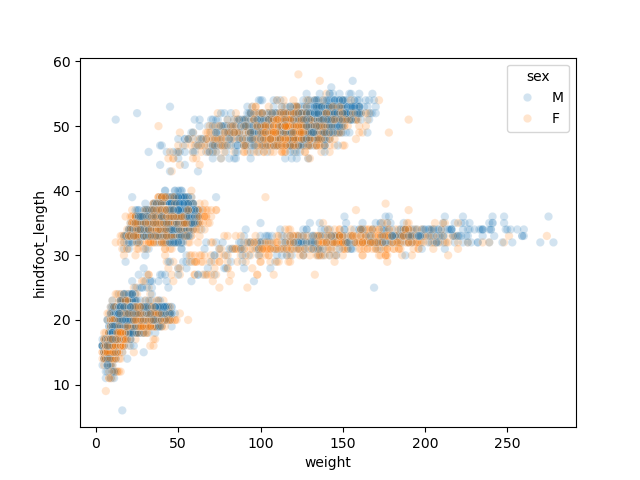
-
Plotly: A tool to explore if you want web-based interactive visualizations where you can hover over data points to get additional information or zoom in to get a closer look. Plots can be displayed in Jupyter notebooks, standalone HTML files, or be part of a web dashboard.
- Load your required libraries into Python and use common nicknames
- Use pandas to load your data –
pd.read_csv()– and to explore it –.head(),.tail(), and.info()methods. - The
.plot()method on your DataFrame is a good plotting starting point. - Matplotlib allows you to customize every aspect of your plot. Start
with the
plt.subplots()function to create a figure object and the number of axes (or subplots) you need. - Export plots to a file using the
.savefig()method.
Content from Exploring and understanding data
Last updated on 2025-10-29 | Edit this page
Overview
Questions
- How can I do exploratory data analysis in Python?
- How do I get help when I am stuck?
- What impact does an object’s type have on what I can do with it?
- How are expressions evaluated and values assigned to variables?
Objectives
- Explore the structure and content of pandas dataframes
- Convert data types and handle missing data
- Interpret error messages and develop strategies to get help with Python
- Trace how Python assigns values to objects
The pandas DataFrame
We just spent quite a bit of time learning how to create
visualisations from the samples data, but we did not talk
much about what samples is. Let’s first load the data
again:
You may remember that we loaded the data into Python with the
pandas.read_csv function. The output of
read_csv is a data frame: a common way of
representing tabular data in a programming language. To be precise,
samples is an object of type
DataFrame. In Python, pretty much everything you work with
is an object of some type. The type function can be used to
tell you the type of any object you pass to it.
OUTPUT
pandas.core.frame.DataFrameThis output tells us that the DataFrame object type is
defined by pandas, i.e. it is a special type of object not included in
the core functionality of Python.
Exploring data in a dataframe
We encountered the plot, head and
tail methods in the previous epsiode. Dataframe objects
carry many other methods, including some that are useful when exploring
a dataset for the first time. Consider the output of
describe:
OUTPUT
month day year plot_id hindfoot_length weight
count 16878.000000 16878.000000 16878.000000 16878.000000 14145.000000 15186.000000
mean 6.382214 15.595805 1983.582119 11.471442 31.982114 53.216647
std 3.411215 8.428180 3.492428 6.865875 10.709841 44.265878
min 1.000000 1.000000 1977.000000 1.000000 6.000000 4.000000
25% 3.000000 9.000000 1981.000000 5.000000 21.000000 24.000000
50% 6.000000 15.000000 1983.000000 11.000000 35.000000 42.000000
75% 9.000000 23.000000 1987.000000 17.000000 37.000000 53.000000
max 12.000000 31.000000 1989.000000 24.000000 70.000000 278.000000These summary statistics give an immediate impression of the distribution of the data. It is always worth performing an initial “sniff test” with these: if there are major issues with the data or its formatting, they may become apparent at this stage.
info provides an overview of the columns included in the
dataframe:
OUTPUT
<class 'pandas.core.frame.DataFrame'>
Index: 16878 entries, 1 to 16878
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 month 16878 non-null int64
1 day 16878 non-null int64
2 year 16878 non-null int64
3 plot_id 16878 non-null int64
4 species_id 16521 non-null object
5 sex 15578 non-null object
6 hindfoot_length 14145 non-null float64
7 weight 15186 non-null float64
8 genus 16521 non-null object
9 species 16521 non-null object
10 taxa 16521 non-null object
11 plot_type 16878 non-null object
dtypes: float64(2), int64(4), object(6)
memory usage: 1.7+ MBWe get quite a bit of useful information here too. First, we are told
that we have a DataFrame of 16878 entries, or rows, and 12
variables, or columns.
Next, we get a bit of information on each variable, including its
column title, a count of the non-null values (that is, values
that are not missing), and something called the dtype of
the column.
Data types
The dtype property of a dataframe column describes the
data type of the values stored in that column. There are three
in the example above:
-
int64: this column contains integer (whole number) values. -
object: this column contains string (non-numeric sequence of characters) values. -
float64: this column contains “floating point” values i.e. numeric values containing a decimal point.
The 64 after int and float
represents the level of precision with which the values in the column
are stored in the computer’s memory. Other types with lower levels of
precision are available for numeric values, e.g. int32 and
float16, which will take up less memory on your system but
limit the size and level of precision of the numbers they can store.
The dtype of a column is important because it determines
the kinds of operation that can be performed on the values in that
column. Let’s work with a couple of the columns independently to
demonstrate this.
The Series object
To work with a single column of a dataframe, we can refer to it by name in two different ways:
or
PYTHON
samples.species_id # this only works if there are no spaces in the column name (note the underscore used here)OUTPUT
record_id
1 NL
2 NL
3 DM
4 DM
5 DM
..
16874 RM
16875 RM
16876 DM
16877 DM
16878 DM
Name: species_id, Length: 16878, dtype: objectTip: use tab completion on column names
Tab completion, where you start typing the name of a variable, function, etc before hitting Tab to auto-complete the rest, also works on column names of a dataframe. Since this tab completion saves time and reduces the chance of including typos, we recommend you use it as frequently as possible.
The result of that operation is a series of data: a
one-dimensional sequence of values that all have the same
dtype (object in this case). Dataframe objects
are collections of the series “glued together” with a shared
index: the column of unique identifiers we associate with each
row. record_id is the index of the series summarised above;
the values carried by the series are NL, DM,
AH, etc (short species identification codes).
If we choose a different column of the dataframe, we get another series with a different data type:
OUTPUT
record_id
1 NaN
2 NaN
3 NaN
4 NaN
5 NaN
...
16874 15.0
16875 9.0
16876 31.0
16877 50.0
16878 42.0
Name: weight, Length: 16878, dtype: float64The data type of the series influences the things that can be done
with/to it. For example, sorting works differently for these two series,
with the numeric values in the weight series sorted from
largest to smallest and the character strings in species_id
sorted alphabetically:
OUTPUT
record_id
9790 4.0
5346 4.0
4052 4.0
9853 4.0
7084 4.0
...
16772 NaN
16777 NaN
16808 NaN
16846 NaN
16860 NaN
Name: weight, Length: 16878, dtype: float64OUTPUT
record_id
12345 AB
9861 AB
10970 AB
10963 AB
5759 AB
...
16453 NaN
16454 NaN
16488 NaN
16489 NaN
16539 NaN
Name: species_id, Length: 16878, dtype: objectThis pattern of behaviour, where the type of an object determines
what can be done with it and influences how it is done, is a defining
characteristic of Python. As you gain more experience with the language,
you will become more familiar with this way of working with data. For
now, as you begin on your learning journey with the language, we
recommend using the type function frequently to make sure
that you know what kind of data/object you are working with, and do not
be afraid to ask for help whenever you are unsure or encounter a
problem.
Aside: Getting Help
You may have already encountered several errors while following the lesson and this is a good time to take a step back and discuss good strategies to get help when something goes wrong.
The built-in help function
Use help to view documentation for an object or
function. For example, if you want to see documentation for the
round function:
OUTPUT
Help on built-in function round in module builtins:
round(number, ndigits=None)
Round a number to a given precision in decimal digits.
The return value is an integer if ndigits is omitted or None. Otherwise
the return value has the same type as the number. ndigits may be negative.The Jupyter Notebook has two ways to get help.
If you are working in Jupyter (Notebook or Lab), the platform offers some additional ways to see documentation/get help:
- Option 1: Type the function name in a cell with a question mark
after it, e.g.
round?. Then run the cell. - Option 2: (Not available on all systems) Place the cursor near where
the function is invoked in a cell (i.e., the function name or its
parameters),
- Hold down Shift, and press Tab.
- Do this several times to expand the information returned.
Understanding error messages
The error messages returned when something goes wrong can be (very)
long but contain information about the problem, which can be very useful
once you know how to interpret it. For example, you might receive a
SyntaxError if you mistyped a line and the resulting code
was invalid:
ERROR
Cell In[129], line 1
name = 'Feng
^
SyntaxError: unterminated string literal (detected at line 1)There are three parts to this error message:
ERROR
Cell In[129], line 1This tells us where the error occured. This is of limited help in
Jupyter, since we know that the error is in the cell we just ran
(Cell In[129]), but the line number can be helpful
especially when the cell is quite long. But when running a larger
program written in Python, perhaps built up from multiple individual
scripts, this can be more useful, e.g.
ERROR
data_visualisation.py, line 42Next, we see a copy of the line where the error was encountered, often annotated with an arrow pointing out exactly where Python thinks the problem is:
ERROR
name = 'Feng
^Python is not exactly right in this case: from context you might be able to guess that the issue is really the lack of a closing quotation mark at the end of the line. But an arrow pointing to the opening quotation mark can give us a push in the right direction. Sometimes Python gets these annotations exactly right. Occasionally, it gets them completely wrong. In the vast majority of cases they are at least somewhat helpful.
Finally, we get the error message itself:
ERROR
SyntaxError: unterminated string literal (detected at line 1)This always begins with a statement of the type of error
encountered: in this case, a SyntaxError. That provides a
broad categorisation for what went wrong. The rest of the message is a
description of exactly what the problem was from Python’s perspective.
Error messages can be loaded with jargon and quite difficult to
understand when you are first starting out. In this example,
unterminated string literal is a technical way of saying
“you opened some quotes, which I think means you were trying to define a
string value, but the quotes did not get closed before the end of the
line.”
It is normal not to understand exactly what these error messages mean the first time you encounter them. Since programming involves making lots of mistakes (for everyone!), you will start to become familiar with many of them over time. As you continue learning, we recommend that you ask others for help: more experienced programmers have made all of these mistakes before you and will probably be better at spotting what has gone wrong. (More on asking for help below.)
Error output can get really long!
Especially when using functions from libraries you have imported into your program, the middle part of the error message (the traceback) can get rather long. For example, what happens if we try to access a column that does not exist in our dataframe?
ERROR
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/miniforge3/envs/carpentries/lib/python3.11/site-packages/pandas/core/indexes/base.py:3812, in Index.get_loc(self, key)
3811 try:
-> 3812 return self._engine.get_loc(casted_key)
3813 except KeyError as err:
File pandas/_libs/index.pyx:167, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/index.pyx:196, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/hashtable_class_helper.pxi:7088, in pandas._libs.hashtable.PyObjectHashTable.get_item()
File pandas/_libs/hashtable_class_helper.pxi:7096, in pandas._libs.hashtable.PyObjectHashTable.get_item()
KeyError: 'wegiht'
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In[131], line 1
----> 1 samples['wegiht']
File ~/miniforge3/envs/carpentries/lib/python3.11/site-packages/pandas/core/frame.py:4107, in DataFrame.__getitem__(self, key)
4105 if self.columns.nlevels > 1:
4106 return self._getitem_multilevel(key)
-> 4107 indexer = self.columns.get_loc(key)
4108 if is_integer(indexer):
4109 indexer = [indexer]
File ~/miniforge3/envs/carpentries/lib/python3.11/site-packages/pandas/core/indexes/base.py:3819, in Index.get_loc(self, key)
3814 if isinstance(casted_key, slice) or (
3815 isinstance(casted_key, abc.Iterable)
3816 and any(isinstance(x, slice) for x in casted_key)
3817 ):
3818 raise InvalidIndexError(key)
-> 3819 raise KeyError(key) from err
3820 except TypeError:
3821 # If we have a listlike key, _check_indexing_error will raise
3822 # InvalidIndexError. Otherwise we fall through and re-raise
3823 # the TypeError.
3824 self._check_indexing_error(key)
KeyError: 'wegiht'(This is still relatively short compared to some errors messages we have seen!)
When you encounter a long error like this one, do not panic! Our advice is to focus on the first couple of lines and the last couple of lines. Everything in the middle (as the name traceback suggests) is retracing steps through the program, identifying where problems were encountered along the way. That information is only really useful to somebody interested in the inner workings of the pandas library, which is well beyond the scope of this lesson! If we ignore everything in the middle, the parts of the error message we want to focus on are:
ERROR
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
[... skipping these parts ...]
KeyError: 'wegiht'This tells us that the problem is the “key”: the value we used to lookup the column in the dataframe. Hopefully, the repetition of the value we provided would be enough to help us realise our mistake.
Other ways to get help
There are several other ways that people often get help when they are stuck with their Python code.
- Search the internet: paste the last line of your error message or
the word “python” and a short description of what you want to do into
your favourite search engine and you will usually find several examples
where other people have encountered the same problem and came looking
for help.
- StackOverflow can be particularly helpful for this: answers to questions are presented as a ranked thread ordered according to how useful other users found them to be.
- Take care: copying and pasting code written by somebody else is risky unless you understand exactly what it is doing!
- ask somebody “in the real world”. If you have a colleague or friend with more expertise in Python than you have, show them the problem you are having and ask them for help.
- Sometimes, the act of articulating your question can help you to identify what is going wrong. This is known as “rubber duck debugging” among programmers.
Generative AI
It is increasingly common for people to use generative AI chatbots such as ChatGPT to get help while coding. You will probably receive some useful guidance by presenting your error message to the chatbot and asking it what went wrong. However, the way this help is provided by the chatbot is different. Answers on StackOverflow have (probably) been given by a human as a direct response to the question asked. But generative AI chatbots, which are based on an advanced statistical model, respond by generating the most likely sequence of text that would follow the prompt they are given.
While responses from generative AI tools can often be helpful, they are not always reliable. These tools sometimes generate plausible but incorrect or misleading information, so (just as with an answer found on the internet) it is essential to verify their accuracy. You need the knowledge and skills to be able to understand these responses, to judge whether or not they are accurate, and to fix any errors in the code it offers you.
In addition to asking for help, programmers can use generative AI tools to generate code from scratch; extend, improve and reorganise existing code; translate code between programming languages; figure out what terms to use in a search of the internet; and more. However, there are drawbacks that you should be aware of.
The models used by these tools have been “trained” on very large volumes of data, much of it taken from the internet, and the responses they produce reflect that training data, and may recapitulate its inaccuracies or biases. The environmental costs (energy and water use) of LLMs are a lot higher than other technologies, both during development (known as training) and when an individual user uses one (also called inference). For more information see the AI Environmental Impact Primer developed by researchers at HuggingFace, an AI hosting platform. Concerns also exist about the way the data for this training was obtained, with questions raised about whether the people developing the LLMs had permission to use it. Other ethical concerns have also been raised, such as reports that workers were exploited during the training process.
We recommend that you avoid getting help from generative AI during the workshop for several reasons:
- For most problems you will encounter at this stage, help and answers can be found among the first results returned by searching the internet.
- The foundational knowledge and skills you will learn in this lesson by writing and fixing your own programs are essential to be able to evaluate the correctness and safety of any code you receive from online help or a generative AI chatbot. If you choose to use these tools in the future, the expertise you gain from learning and practising these fundamentals on your own will help you use them more effectively.
- As you start out with programming, the mistakes you make will be the kinds that have also been made – and overcome! – by everybody else who learned to program before you. Since these mistakes and the questions you are likely to have at this stage are common, they are also better represented than other, more specialised problems and tasks in the data that was used to train generative AI tools. This means that a generative AI chatbot is more likely to produce accurate responses to questions that novices ask, which could give you a false impression of how reliable they will be when you are ready to do things that are more advanced.
Data input within Python
Although it is more common (and faster) to input data in another format e.g. a spreadsheet and read it in, Series and DataFrame objects can be created directly within Python. Before we can make a new Series, we need to learn about another type of data in Python: the list.
Lists
Lists are one of the standard data structures built into
Python. A data structure is an object that contains more than one piece
of information. (DataFrames and Series are also data structures.) The
list is designed to contain multiple values in an ordered sequence: they
are a great choice if you want to build up and modify a collection of
values over time and/or handle each of those values one at a time. We
can create a new list in Python by capturing the values we want it to
store inside square brackets []:
OUTPUT
[2020, 2025, 2010]New values can be added to the end of a list with the
append method:
OUTPUT
[2020, 2025, 2010, 2015]Exploring list methods
The append method allows us to add a value to the end of
a list but how could we insert a new value into a given position
instead? Applying what you have learned about how to find out the
methods that an object has, can you figure out how to place the value
2019 into the third position in years_list
(shifting the values after it up one more position)? Recall that the
indexing used to specify positions in a sequence begins at 0 in
Python.
Using tab completion, the help function, or looking up
the documentation online, we can discover the insert method
and learn how it works. insert takes two arguments: the
position for the new list entry and the value to be placed in that
position:
OUTPUT
[2020, 2025, 2019, 2010, 2015]Among many other methods is sort, which can be used to
sort the values in the list:
OUTPUT
[2010, 2015, 2019, 2020, 2025]The easiest way to create a new Series is with a list:
OUTPUT
0 2010
1 2015
2 2019
3 2020
4 2025
dtype: int64With the data in a Series, we can no longer do some of the things we were able to do with the list, such as adding new values. But we do gain access to some new possibilities, which can be very helpful. For example, if we wanted to increase all of the values by 1000, this would be easy with a Series but more complicated with a list:
OUTPUT
0 3010
1 3015
2 3019
3 3020
4 3025
dtype: int64ERROR
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[126], line 1
----> 1 years_list + 1000
TypeError: can only concatenate list (not "int") to listThis illustrates an important principle of Python: different data structures are suitable for different “modes” of working with data. It can be helpful to work with a list when building up an initial set of data from scratch, but when you are ready to begin operating on that dataset as a whole (performing calculations with it, visualising it, etc), you will be rewarded for switching to a more specialised datatype like a Series or DataFrame from pandas.
Unexpected data types
Operations like the addition of 1000 we performed on
years_series work because pandas knows how to add a number
to the integer values in the series. That behaviour is determind by the
dtype of the series, which makes that dtype
really important for how you want to work with your data. Let’s explore
how the dtype is chosen. Returning to the
years_series object we created above:
OUTPUT
0 2010
1 2015
2 2019
3 2020
4 2025
dtype: int64The dtype: int64 was determined automatically based on
the values passed in. But what if the values provided are of several
different types?
OUTPUT
0 2.0
1 3.0
2 5.5
3 6.0
4 8.0
dtype: float64Pandas assigns a dtype that allows it to account for all
of the values it is given, converting some values to another
dtype if needed, in a process called coercion. In
the case above, all of the integer values were coerced to floating point
numbers to account for the 5.5.
In practice, it is much more common to read data from elsewhere
(e.g. with read_csv) than to enter it manually within
Python. When reading data from a file, pandas tries to guess the
appropriate dtype to assign to each column (series) of the
dataframe. This is usually very helpful but the process is sensitive to
inconsistencies and data entry errors in the input: a stray character in
one cell can cause an entire column to be coerced to a different
dtype than you might have wanted.
For example, if the raw data includes a symbol added by a typo
mistake (= instead of -):
name,latitude,longitude
Superior,47.7,-87.5
Victoria,-1.0,33.0
Tanganyika,=6.0,29.3We see a non-numeric dtype for the latitude column
(object) when we load the data into a dataframe.
OUTPUT
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3 entries, 0 to 2
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 name 3 non-null object
1 latitude 3 non-null object
2 longitude 3 non-null float64
dtypes: float64(1), object(2)
memory usage: 200.0+ bytesIt is a good idea to run the info method on a new
dataframe after you have loaded data for the first time: if one or more
of the columns has a different dtype than you expected,
this may be a signal that you need to clean up the raw data.
Recasting
A column can be manually coerced (or recast) into a
different dtype, provided that pandas knows how to handle
that conversion. For example, the integer values in the
plot_id column of our dataframe can be converted to
floating point numbers:
OUTPUT
record_id
record_id
1 2.0
2 3.0
3 2.0
4 7.0
5 3.0
...
16874 16.0
16875 5.0
16876 4.0
16877 11.0
16878 8.0
Name: plot_id, Length: 16878, dtype: float64But the string values of species_id cannot be converted
to numeric data:
ERROR
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[101], line 1
----> 1 samples.species_id = samples.species_id.astype('int64')
File ~/miniforge3/envs/carpentries/lib/python3.11/site-packages/pandas/core/generic.py:6662, in NDFrame.astype(self, dtype, copy, errors)
6656 results = [
6657 ser.astype(dtype, copy=copy, errors=errors) for _, ser in self.items()
6658 ]
6660 else:
6661 # else, only a single dtype is given
-> 6662 new_data = self._mgr.astype(dtype=dtype, copy=copy, errors=errors)
6663 res = self._constructor_from_mgr(new_data, axes=new_data.axes)
6664 return res.__finalize__(self, method="astype")
[... a lot more lines of traceback ...]
File ~/miniforge3/envs/carpentries/lib/python3.11/site-packages/pandas/core/dtypes/astype.py:133, in _astype_nansafe(arr, dtype, copy, skipna)
129 raise ValueError(msg)
131 if copy or arr.dtype == object or dtype == object:
132 # Explicit copy, or required since NumPy can't view from / to object.
--> 133 return arr.astype(dtype, copy=True)
135 return arr.astype(dtype, copy=copy)
ValueError: invalid literal for int() with base 10: 'NL'Changing Types
- Convert the values in the column
plot_idback to integers. - Now try converting
weightto an integer. What goes wrong here? What is pandas telling you? We will talk about some solutions to this later.
OUTPUT
record_id
1 2
2 3
3 2
4 7
5 3
..
16874 16
16875 5
16876 4
16877 11
16878 8
Name: plot_id, Length: 16878, dtype: int64ERROR
pandas.errors.IntCastingNaNError: Cannot convert non-finite values (NA or inf) to integerPandas cannot convert types from float to int if the column contains missing values.
Missing Data
In addition to data entry errors, it is common to encounter missing values in large volumns of data. A value may be missing because it was not possible to make a complete observation, because data was lost, or for any number of other reasons. It is important to consider missing values while processing data because they can influence downstream analysis – that is, data analysis that will be done later – in unwanted ways when not handled correctly.
Depending on the dtype of the column/series, missing
values may appear as NaN (“Not a Number”),
NA, <NA>, or NaT (“Not
a Time”). You may have noticed some during our initial exploration
of the dataframe. (Note the NaN values in the first five
rows of the weight column below.)
OUTPUT
month day year plot_id species_id sex hindfoot_length weight genus species taxa plot_type
record_id
1 7 16 1977 2 NL M 32.0 NaN Neotoma albigula Rodent Control
2 7 16 1977 3 NL M 33.0 NaN Neotoma albigula Rodent Long-term Krat Exclosure
3 7 16 1977 2 DM F 37.0 NaN Dipodomys merriami Rodent Control
4 7 16 1977 7 DM M 36.0 NaN Dipodomys merriami Rodent Rodent Exclosure
5 7 16 1977 3 DM M 35.0 NaN Dipodomys merriami Rodent Long-term Krat ExclosureThe output of the info method includes a count of the
non-null values – that is, the values that are not
missing – for each column:
OUTPUT
<class 'pandas.core.frame.DataFrame'>
Index: 16878 entries, 1 to 16878
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 month 16878 non-null int64
1 day 16878 non-null int64
2 year 16878 non-null int64
3 plot_id 16878 non-null int64
4 species_id 16521 non-null object
5 sex 15578 non-null object
6 hindfoot_length 14145 non-null float64
7 weight 15186 non-null float64
8 genus 16521 non-null object
9 species 16521 non-null object
10 taxa 16521 non-null object
11 plot_type 16878 non-null object
dtypes: float64(2), int64(4), object(6)
memory usage: 1.7+ MBFrom this output we can tell that almost 1700 weight measurements and more than 2700 hindfoot length measurements are missing. Many of the other columns also have missing values.
The ouput above demonstrates that pandas can distinguish these
NaN values from the actual data and indeed they will be
ignored for some tasks, such as calculation of the summary statistics
provided by describe.
OUTPUT
month day year plot_id hindfoot_length weight
count 16878.000000 16878.000000 16878.000000 16878.000000 14145.000000 15186.000000
mean 6.382214 15.595805 1983.582119 11.471442 31.982114 53.216647
std 3.411215 8.428180 3.492428 6.865875 10.709841 44.265878
min 1.000000 1.000000 1977.000000 1.000000 6.000000 4.000000
25% 3.000000 9.000000 1981.000000 5.000000 21.000000 24.000000
50% 6.000000 15.000000 1983.000000 11.000000 35.000000 42.000000
75% 9.000000 23.000000 1987.000000 17.000000 37.000000 53.000000
max 12.000000 31.000000 1989.000000 24.000000 70.000000 278.000000In some circumstances, like the recasting we attempted in the previous exercise, the missing values can cause trouble. It is up to us to decide how best to handle those missing values. We could remove the rows containing missing data, accepting the loss of all data for that observation:
OUTPUT
month day year plot_id species_id sex hindfoot_length weight genus species taxa plot_type
record_id
63 8 19 1977 3 DM M 35.0 40.0 Dipodomys merriami Rodent Long-term Krat Exclosure
64 8 19 1977 7 DM M 37.0 48.0 Dipodomys merriami Rodent Rodent Exclosure
65 8 19 1977 4 DM F 34.0 29.0 Dipodomys merriami Rodent Control
66 8 19 1977 4 DM F 35.0 46.0 Dipodomys merriami Rodent Control
67 8 19 1977 7 DM M 35.0 36.0 Dipodomys merriami Rodent Rodent ExclosureBut we should take note that this removes more than 3000 rows from the dataframe!
OUTPUT
16878OUTPUT
13773Instead, we could fill all of the missing values with
something else. For example, let’s make a copy of the
samples dataframe then populate the missing values in the
weight column of that copy with the mean of all the
non-missing weights. There are a few parts to that operation, which are
tackled one at a time below.
PYTHON
mean_weight = samples['weight'].mean() # the 'mean' method calculates the mean of the non-null values in the column
df1 = samples.copy() # making a copy to work with so that we do not edit our original data
df1['weight'] = df1['weight'].fillna(mean_weight) # the 'fillna' method fills all missing values with the provided value
df1.head()OUTPUT
month day year plot_id species_id sex hindfoot_length weight genus species taxa plot_type
record_id
1 7 16 1977 2 NL M 32.0 53.216647 Neotoma albigula Rodent Control
2 7 16 1977 3 NL M 33.0 53.216647 Neotoma albigula Rodent Long-term Krat Exclosure
3 7 16 1977 2 DM F 37.0 53.216647 Dipodomys merriami Rodent Control
4 7 16 1977 7 DM M 36.0 53.216647 Dipodomys merriami Rodent Rodent Exclosure
5 7 16 1977 3 DM M 35.0 53.216647 Dipodomys merriami Rodent Long-term Krat ExclosureThe choice to fill in these missing values rather than removing the rows that contain them can have implications for the result of your analysis. It is important to consider your approach carefully. Think about how the data will be used and how these values will impact the scientific conclusions made from the analysis. pandas gives us all of the tools that we need to account for these issues. But we need to be cautious about how the decisions that we make impact scientific results.
Assignment, evaluation, and mutability
Stepping away from dataframes for a moment, the time has come to explore the behaviour of Python a little more.
Understanding what’s going on here will help you avoid a lot of
confusion when working in Python. When we assign something to a
variable, the first thing that happens is the righthand side gets
evaluated. So when we first ran the line y = x*3,
x*3 first gets evaluated to the value of 6, and this gets
assigned to y. The variables x and
y are independent objects, so when we change the value of
x to 10, y is unaffected. This behaviour may
be different to what you are used to, e.g. from experience working with
data in spreadsheets where cells can be linked such that modifying the
value in one cell triggers changes in others.
Multiple evaluations can take place in a single line of Python code and learning to trace the order and impact of these evaluations is a key skill.
OUTPUT
1.0In the example above, x/y is evaluated first before the
result is subtracted from 3 and the final calculated value is assigned
to z. (The brackets () are not needed in the
calculation above but are included to make the order of evaluation
clearer.) Python makes each evaluation as it needs to in order to
proceed with the next, before assigning the final result to the variable
on the lefthand side of the = operator.
This means that we could have filled the missing values in the weight column of our dataframe copy in a single line:
First, the mean weight is calculated
(df1['weight'].mean() is evaluated). Then the result of
that evaluation is passed into fillna and the result of the
filling operation
(df1['weight'].fillna(<RESULT OF PREVIOUS>)) is
assigned to df1['weight'].
Variable naming
You are going to name a lot of variables in Python! There are some rules you have to stick to when doing so, as well as recommendations that will make your life easier.
- Make names clear without being too long
-
wkgis probably too short. -
weight_in_kilogramsis probably too long. -
weight_kgis good.
-
- Names cannot begin with a number.
- Names cannot contain spaces; use underscores instead.
- Names are case sensitive:
weight_kgis a different name fromWeight_kg. Avoid uppercase characters at the beginning of variable names. - Names cannot contain most non-letter characters:
+&-/*.etc. - Two common formats of variable name are
snake_caseandcamelCase. A third “case” of naming convention,kebab-case, is not allowed in Python (see the rule above). - Aim to be consistent in how you name things within your projects. It is easier to follow an established style guide, such as Google’s, than to come up with your own.
Exercise; good variable names
Identify at least one good variable name and at least one variable name that could be improved in this episode. Refer to the rules and recommendations listed above to suggest how these variable names could be better.
mean_weight and samples are examples of
reasonably good variable names: they are relatively short yet
descriptive.
df2 is not descriptive enough and could be potentially
confusing if encountered by somebody else/ourselves in a few weeks’
time. The name could be improved by making it more descriptive,
e.g. samples_duplicate.
Mutability
Why did we need to use the copy method to duplicate the
dataframe above if variables are not linked to each other? Why not
assign a new variable with the value of the existing dataframe
object?
This gets to mutablity: a feature of Python that has caused headaches for many novices in the past! In the interests of memory management, Python avoids making copies of objects unless it has to. Some types of objects are immutable, meaning that their value cannot be modified once set. Immutable object types we have already encountered include strings, integers, and floats. If we want to adjust the value of an integer variable, we must explicitly overwrite it.
Other types of object are mutable, meaning that their value
can be changed “in-place” without needing to be explictly overwritten.
This includes lists and pandas DataFrame
objects, which can be reordered etc. after they are
created.
When a new variable is assigned the value of an existing immutable object, Python duplicates the value and assigns it to the new variable.
a = 3.5 # new float object, called "a"
b = a # another new float object, called "b", which also has the value 3.5When a new variable is assigned the value of an existing mutable object, Python makes a new “pointer” towards the value of the existing object instead of duplicating it.
some_species = ['NL', 'DM', 'PF', 'PE', 'DS'] # new list object, called "some_species"
some_more_species = some_species # another name for the same list objectThis can have unintended consequences and lead to much confusion!
some_more_species[2] = 'CV'
some_speciesOUTPUT
['NL', 'DM', 'CV', 'PE', 'DS']As you can see here, the “PV” value was replaced by “CV” in both
lists, even if we didn’t intend to make the change in the
some_species list.
This takes practice and time to get used to. The key thing to
remember is that you should use the copy method to
make a copy of your dataframes to avoid accidentally modifying
the data in the original.
Groups in Pandas
We often want to calculate summary statistics grouped by subsets or attributes within fields of our data. For example, we might want to calculate the average weight of all individuals per site.
We can calculate basic statistics for all records in a single column using the syntax below:
gives output
PYTHON
count 15186.000000
mean 53.216647
std 44.265878
min 4.000000
25% 24.000000
50% 42.000000
75% 53.000000
max 278.000000
Name: weight, dtype: float64We can also extract one specific metric if we wish:
PYTHON
samples['weight'].min()
samples['weight'].max()
samples['weight'].mean()
samples['weight'].std()
samples['weight'].count()But if we want to summarize by one or more variables, for example
sex, we can use Pandas’ .groupby method.
Once we’ve created a groupby DataFrame, we can quickly calculate summary
statistics by a group of our choice.
The pandas function describe will
return descriptive stats including: mean, median, max, min, std and
count for a particular column in the data. Pandas’ describe
function will only return summary values for columns containing numeric
data.
PYTHON
# Summary statistics for all numeric columns by sex
grouped_data.describe()
# Provide the mean for each numeric column by sex
grouped_data.mean(numeric_only=True)OUTPUT
record_id month day year plot_id \
sex
F 8371.632960 6.475266 15.411998 1983.520361 11.418147
M 8553.106416 6.306295 15.763317 1983.679177 11.248305
hindfoot_length weight
sex
F 31.83258 53.114471
M 32.13352 53.164464
The groupby command is powerful in that it allows us to
quickly generate summary stats.
Challenge - Summary Data
- How many recorded individuals are female
Fand how many maleM? - What happens when you group by two columns using the following syntax and then calculate mean values?
grouped_data2 = samples.groupby(['plot_id', 'sex'])grouped_data2.mean(numeric_only=True)
- Summarize weight values for each site in your data. HINT: you can
use the following syntax to only create summary statistics for one
column in your data.
by_site['weight'].describe()
- The first column of output from
grouped_data.describe()(count) tells us that the data contains 15690 records for female individuals and 17348 records for male individuals.- Note that these two numbers do not sum to 35549, the total number of
rows we know to be in the
samplesDataFrame. Why do you think some records were excluded from the grouping?
- Note that these two numbers do not sum to 35549, the total number of
rows we know to be in the
- Calling the
mean()method on data grouped by these two columns calculates and returns the mean value for each combination of plot and sex.- Note that the mean is not meaningful for some variables, e.g. day,
month, and year. You can specify particular columns and particular
summary statistics using the
agg()method (short for aggregate), e.g. to obtain the last survey year, median foot-length and mean weight for each plot/sex combination:
- Note that the mean is not meaningful for some variables, e.g. day,
month, and year. You can specify particular columns and particular
summary statistics using the
PYTHON
samples.groupby(['plot_id', 'sex']).agg({"year": 'max',
"hindfoot_length": 'median',
"weight": 'mean'})samples.groupby(['plot_id'])['weight'].describe()
OUTPUT
count mean std min 25% 50% 75% max
plot_id
1 909.0 65.974697 45.807013 4.0 39.0 46.0 99.00 223.0
2 962.0 59.911642 50.234865 5.0 31.0 44.0 55.00 278.0
3 641.0 38.338534 50.623079 4.0 11.0 20.0 34.00 250.0
4 828.0 62.647343 41.208190 4.0 37.0 45.0 102.00 200.0
5 788.0 47.864213 36.739691 5.0 28.0 42.0 50.00 248.0
6 686.0 49.180758 36.620356 5.0 25.0 42.0 52.00 243.0
7 257.0 25.101167 31.649778 4.0 11.0 19.0 24.00 235.0
8 736.0 64.593750 43.420011 5.0 39.0 48.0 102.25 178.0
9 893.0 65.346025 41.928699 6.0 40.0 48.0 99.00 275.0
10 159.0 21.188679 25.744403 4.0 10.0 12.0 24.50 237.0
11 905.0 50.260773 37.034074 5.0 29.0 42.0 49.00 212.0
12 1086.0 55.978821 45.675559 7.0 31.0 44.0 53.00 264.0
13 647.0 56.316847 42.464628 5.0 30.5 44.0 54.00 241.0
14 798.0 52.909774 33.993126 5.0 38.0 45.0 51.00 216.0
15 357.0 35.011204 47.396960 4.0 10.0 19.0 33.00 259.0
16 232.0 26.185345 22.040403 4.0 11.0 20.0 40.00 158.0
17 788.0 59.426396 44.751988 4.0 30.0 45.0 61.25 216.0
18 690.0 56.000000 44.368296 5.0 29.0 42.0 53.00 256.0
19 369.0 19.059621 15.320905 4.0 9.0 15.0 23.00 139.0
20 662.0 65.531722 53.234713 6.0 30.0 44.0 110.75 223.0
21 342.0 24.964912 32.230001 4.0 8.0 12.0 27.00 190.0
22 611.0 70.163666 45.955603 5.0 37.0 48.0 118.00 212.0
23 209.0 21.502392 19.647158 4.0 10.0 16.0 24.00 131.0
24 631.0 50.123613 47.017531 4.0 23.0 40.0 47.00 251.0Quick & Easy Plotting Data Using Pandas
We can plot our summary stats using Pandas, too.
We can also look at how many animals were captured in each site:
PYTHON
total_count = samples.groupby('plot_id')['record_id'].nunique()
# Let's plot that too
total_count.plot(kind='bar')Challenge - Plots
- Create a plot of average weight across all species per site.
- Create a plot of total males versus total females for the entire dataset.
samples.groupby('plot_id')["weight"].mean().plot(kind='bar')
samples.groupby('sex')["record_id"].count().plot(kind='bar')
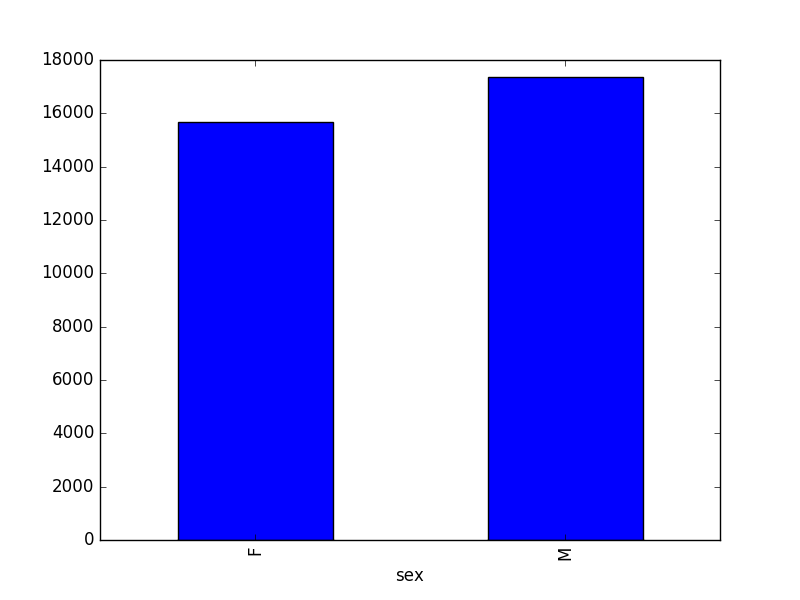
- pandas DataFrames carry many methods that can help you explore the properties and distribution of data.
- Using the
helpfunction, reading error messages, and asking for help are all good strategies when things go wrong. - The type of an object determines what kinds of operations you can perform on and with it.
- Python evaluates expressions in a line one by one before assigning the final result to a variable.
Content from Indexing, Slicing and Subsetting DataFrames
Last updated on 2025-10-29 | Edit this page
Overview
Questions
- How can I access specific data within my data set?
- How can Python and Pandas help me to analyse my data?
Objectives
- Describe what 0-based indexing is.
- Manipulate and extract data using column headings and index locations.
- Employ slicing to select sets of data from a DataFrame.
- Employ label and integer-based indexing to select ranges of data in a dataframe.
- Reassign values within subsets of a DataFrame.
- Create a copy of a DataFrame.
- Query / select a subset of data using a set of criteria using the
following operators:
==,!=,>,<,>=,<=. - Locate subsets of data using masks.
- Describe BOOLEAN objects in Python and manipulate data using BOOLEANs.
In the first episode of this lesson, we read a CSV file into a pandas’ DataFrame. We learned how to:
- save a DataFrame to a named object,
- create plots based on the data we loaded into pandas
In this lesson, we will explore ways to access different parts of the data using:
- indexing,
- slicing, and
- subsetting
Loading our data
We will continue to use the samples dataset that we worked with in the last episode. Let’s reopen and read in the data again:
Indexing and Slicing in Python
We often want to work with subsets of a DataFrame object. There are different ways to accomplish this including: using labels (column headings), numeric ranges, or specific x,y index locations.
Selecting data using Labels (Column Headings)
We use square brackets [] to select a subset of a Python
object. For example, we can select all data from a column named
species_id from the samples DataFrame by name.
There are two ways to do this:
PYTHON
# TIP: use the .head() method we saw earlier to make output shorter
# Method 1: select a 'subset' of the data using the column name
samples['species_id']
# Method 2: use the column name as an 'attribute'; gives the same output
samples.species_idWe can pass a list of column names too, as an index to select columns in that order. This is useful when we need to reorganize our data.
NOTE: If a column name is not contained in the DataFrame, an exception (error) will be raised.
PYTHON
# Select the species and plot columns from the DataFrame
samples[['species_id', 'plot_id']]
# What happens when you flip the order?
samples[['plot_id', 'species_id']]
# What happens if you ask for a column that doesn't exist?
samples['speciess']Python tells us what type of error it is in the traceback, at the
bottom it says KeyError: 'speciess' which means that
speciess is not a valid column name (nor a valid key in the
related Python data type dictionary).
Reminder
The Python language and its modules (such as Pandas) define reserved
words that should not be used as identifiers when assigning objects and
variable names. Examples of reserved words in Python include Boolean
values True and False, operators
and, or, and not, among others.
The full list of reserved words for Python version 3 is provided at https://docs.python.org/3/reference/lexical_analysis.html#identifiers.
When naming objects and variables, it’s also important to avoid using
the names of built-in data structures and methods. For example, a
list is a built-in data type. It is possible to use the word
‘list’ as an identifier for a new object, for example
list = ['apples', 'oranges', 'bananas']. However, you would
then be unable to create an empty list using list() or
convert a tuple to a list using list(sometuple).
Extracting Range based Subsets: Slicing
Reminder
Python uses 0-based indexing.
Let’s remind ourselves that Python uses 0-based indexing. This means
that the first element in an object is located at position
0. This is different from other tools like R and Matlab
that index elements within objects starting at 1.
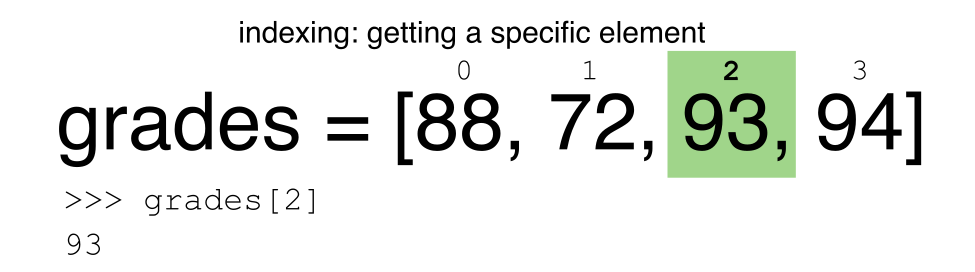
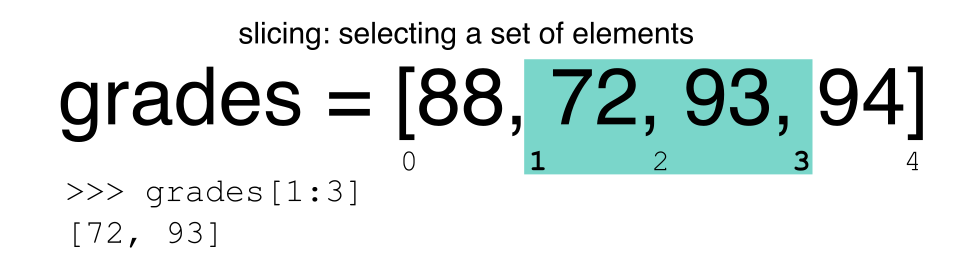
a[0]returns1, as Python starts with element 0 (this may be different from what you have previously experience with other languages e.g. MATLAB and R)a[5]raises anIndexErrorThe error is raised because the list
ahas no element with index 5: it has only five entries, indexed from 0 to 4.-
a[len(a)]also raises anIndexError.len(a)returns5, makinga[len(a)]equivalent toa[5]. To retreive the final element of a list, us the index-1, e.g.OUTPUT
5
Slicing Subsets of Rows in Python
Slicing using the [] operator selects a set of rows
and/or columns from a DataFrame. To slice out a set of rows, you use the
following syntax: data[start:stop]. When slicing in pandas
the start bound is included in the output. The stop bound is one step
BEYOND the row you want to select. So if you want to select rows 0, 1
and 2 your code would look like this:
The stop bound in Python is different from what you might be used to in languages like Matlab and R.
Slicing Subsets of Rows and Columns in Python
We just learned we can use square brackets to slice our DataFrame and
select columns or rows. And this is useful overall in Python for
selecting a subset of items from a data structure. However, the pandas
library recommends to use .loc[] and .iloc[]
instead. With bot of these we can select specific ranges of our data in
both the row and column directions, using either label or integer-based
indexing.
locis primarily a label based indexing where you can refer to rows and columns by their name. E.g., column ‘month’. Note that integers may be used, but they are interpreted as a label.ilocis primarily an integer based indexing counting from 0. That is, you specify rows and columns using a number. Thus, the first row is row 0, the second column is column 1, etc.
To select a subset of rows and columns from our
DataFrame, we can use the loc method. For example, we can
select the first three rows, and the month, day and year columns, like
this:
which gives the output
OUTPUT
month day year
0 7 16 1977
1 7 16 1977
2 7 16 1977
3 7 16 1977Let’s explore some other ways to index and select subsets of data:
PYTHON
# Select all columns for rows of index values 0 and 10
samples.loc[[0, 10], :]
# What does this do?
samples.loc[0, ['species_id', 'plot_id', 'weight']]
# What happens when you type the code below?
samples.loc[[0, 10, 35549], :]NOTE: Labels must be found in the DataFrame or you
will get a KeyError.
We can also select a specific data value using a row and column
location within the DataFrame and iloc indexing:
In this iloc example,
gives the output
OUTPUT
'F'Remember that Python indexing begins at 0. So, the index location [2, 6] selects the element that is 3 rows down and 7 columns over in the DataFrame.
Challenge - Range
- What happens when you execute:
samples[0:1]samples[0]samples[:4]samples[:-1]
- What happens when you call:
samples.iloc[0:1]samples.iloc[0]samples.iloc[:4, :]samples.iloc[0:4, 1:4]samples.loc[0:4, 1:4]
- How are the last two commands different?
-
-
samples[0:3]returns the first three rows of the DataFrame:
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 0 1 7 16 1977 2 NL M 32.0 NaN 1 2 7 16 1977 3 NL M 33.0 NaN 2 3 7 16 1977 2 DM F 37.0 NaN-
samples[0]results in a ‘KeyError’, since direct indexing of a row is redundant withiloc. -
samples[0:1]can be used to obtain only the first row. -
samples[:5]slices from the first row to the fifth:
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length weight 0 1 7 16 1977 2 NL M 32.0 NaN 1 2 7 16 1977 3 NL M 33.0 NaN 2 3 7 16 1977 2 DM F 37.0 NaN 3 4 7 16 1977 7 DM M 36.0 NaN 4 5 7 16 1977 3 DM M 35.0 NaN-
samples[:-1]provides everything except the final row of the DataFrame. You can use negative index numbers to count backwards from the last entry.
-
-
samples.iloc[0:1]returns the first row -
samples.iloc[0]returns the first row as a named list -
samples.iloc[:4, :]returns all columns of the first four rows -
samples.iloc[0:4, 1:4]selects specified columns of the first four rows -
samples.loc[0:4, 1:4]results in a ‘TypeError’ - see below.
-
- While
ilocuses integers as indices and slices accordingly,locworks with labels. It is like accessing values from a dictionary, asking for the key names. Column names 1:4 do not exist, so the call tolocabove results in an error. Check also the difference betweensamples.loc[0:4]andsamples.iloc[0:4].
Subsetting Data using Criteria
We can also select a subset of our data using criteria. For example, we can select all rows that have a year value of 2002:
Which produces the following output:
PYTHON
record_id month day year plot_id species_id sex hindfoot_length \
9789 9790 1 19 1985 16 RM F 16.0
9790 9791 1 19 1985 17 OT F 20.0
9791 9792 1 19 1985 6 DO M 35.0
9792 9793 1 19 1985 12 DO F 35.0
9793 9794 1 19 1985 24 RM M 16.0
... ... ... ... ... ... ... ... ...
11222 11223 12 8 1985 4 DM M 36.0
11223 11224 12 8 1985 11 DM M 37.0
11224 11225 12 8 1985 7 PE M 20.0
11225 11226 12 8 1985 1 DM M 38.0
11226 11227 12 8 1985 15 NaN NaN NaN
weight genus species taxa plot_type
9789 4.0 Reithrodontomys megalotis Rodent Rodent Exclosure
9790 16.0 Onychomys torridus Rodent Control
9791 48.0 Dipodomys ordii Rodent Short-term Krat Exclosure
9792 40.0 Dipodomys ordii Rodent Control
9793 4.0 Reithrodontomys megalotis Rodent Rodent Exclosure
... ... ... ... ... ...
11222 40.0 Dipodomys merriami Rodent Control
11223 49.0 Dipodomys merriami Rodent Control
11224 18.0 Peromyscus eremicus Rodent Rodent Exclosure
11225 47.0 Dipodomys merriami Rodent Spectab exclosure
11226 NaN NaN NaN NaN Long-term Krat Exclosure
[1438 rows x 13 columns]Or we can select all rows that do not contain the year 2002:
We can define sets of criteria too:
Python Syntax Cheat Sheet
We can use the syntax below when querying data by criteria from a DataFrame. Experiment with selecting various subsets of the “samples” data.
- Equals:
== - Not equals:
!= - Greater than:
> - Less than:
< - Greater than or equal to:
>= - Less than or equal to:
<=
Challenge - Queries
Select a subset of rows in the
samplesDataFrame that contain data from the year 1999 and that contain weight values less than or equal to 8. How many rows did you end up with? What did your neighbor get?-
You can use the
isincommand in Python to query a DataFrame based upon a list of values as follows:Use the
isinfunction to find all plots that contain the particular speciesNLandDMin the “samples” DataFrame. How many records contain these values? Experiment with other queries. Create a query that finds all rows with a weight value greater than or equal to 0.
The
~symbol in Python can be used to return the OPPOSITE of the selection that you specify. It is equivalent to is not in. Write a query that selects all rows with sex NOT equal to ‘M’ or ‘F’ in the “samples” data.
-
If you are only interested in how many rows meet the criteria, the sum of
Truevalues could be used instead:OUTPUT
63 -
For example, using
NLandDM:OUTPUT
array([ 2, 3, 7, 1, 8, 6, 4, 14, 15, 13, 9, 11, 10, 17, 20, 23, 21, 18, 22, 19, 12, 5, 16, 24])OUTPUT
(24,) samples.loc[samples["weight"] >= 0, :]
Using masks to identify a specific condition
A mask can be useful to locate where a particular
subset of values exist or don’t exist - for example, NaN, or “Not a
Number” values. To understand masks, we also need to understand
BOOLEAN objects in Python.
Boolean values include True or False. For
example,
When we ask Python whether x is greater than 5, it
returns False. This is Python’s way to say “No”. Indeed,
the value of x is 5, and 5 is not greater than 5.
To create a boolean mask:
- Set the True / False criteria
(e.g.
values > 5 = True) - Python will then assess each value in the object to determine whether the value meets the criteria (True) or not (False).
- Python creates an output object that is the same shape as the
original object, but with a
TrueorFalsevalue for each index location.
Let’s try this out. Let’s identify all locations in the survey data
that have null (missing or NaN) data values. We can use the
isna method to do this. The isna method will
compare each cell with a null value. If an element has a null value, it
will be assigned a value of True in the output object.
A snippet of the output is below:
PYTHON
record_id month day year plot_id species_id sex \
0 False False False False False False False
1 False False False False False False False
2 False False False False False False False
3 False False False False False False False
4 False False False False False False False
... ... ... ... ... ... ... ...
16873 False False False False False False False
16874 False False False False False False False
16875 False False False False False False False
16876 False False False False False False False
16877 False False False False False False False
hindfoot_length weight genus species taxa plot_type
0 False True False False False False
1 False True False False False False
2 False True False False False False
3 False True False False False False
4 False True False False False False
... ... ... ... ... ... ...
16873 False False False False False False
16874 False False False False False False
16875 False False False False False False
16876 False False False False False False
16877 False False False False False False
[16878 rows x 13 columns]To select the rows where there are null values, we can use the mask as an index to subset our data as follows:
PYTHON
# To select just the rows with NaN values, we can use the 'any()' method
samples.loc[samples.isna().any(axis=1), :]We can run isna on a particular column too. What does
the code below do?
PYTHON
# What does this do?
empty_weights = samples.loc[samples["weight"].isna(), "weight"]
print(empty_weights)Let’s take a minute to look at the statement above. We are using the
Boolean object samples["weight"].isna() as an index to
samples. We are asking Python to select rows that have a
NaN value of weight.
Challenge - Putting it all together
Create a new DataFrame that only contains observations with sex values that are not female or male. Print the number of rows in this new DataFrame. Verify the result by comparing the number of rows in the new DataFrame with the number of rows in the samples DataFrame where sex is null.
- In Python, portions of data can be accessed using indices, slices, column headings, and condition-based subsetting.
- Python uses 0-based indexing, in which the first element in a list, tuple or any other data structure has an index of 0.
- Pandas enables common data exploration steps such as data indexing, slicing and conditional subsetting.
Content from Combining DataFrames
Last updated on 2025-10-29 | Edit this page
Overview
Questions
- Can I work with data from multiple sources?
- How can I combine data from different data sets?
Objectives
- Combine data from multiple files into a single DataFrame using merge and concat.
- Combine two DataFrames using a unique ID found in both DataFrames.
- Employ
to_csvto export a DataFrame in CSV format. - Join DataFrames using common fields (join keys).
In many “real world” situations, the data that we want to use come in
multiple files. We often need to combine these files into a single
DataFrame to analyze the data. The pandas package provides various
methods for combining DataFrames including merge and
concat.
To work through the examples below, we first need to load the species and surveys files into pandas DataFrames.:
OUTPUT
record_id month day year plot species sex hindfoot_length weight
0 1 7 16 1977 2 NA M 32 NaN
1 2 7 16 1977 3 NA M 33 NaN
2 3 7 16 1977 2 DM F 37 NaN
3 4 7 16 1977 7 DM M 36 NaN
4 5 7 16 1977 3 DM M 35 NaN
... ... ... ... ... ... ... ... ... ...
35544 35545 12 31 2002 15 AH NaN NaN NaN
35545 35546 12 31 2002 15 AH NaN NaN NaN
35546 35547 12 31 2002 10 RM F 15 14
35547 35548 12 31 2002 7 DO M 36 51
35548 35549 12 31 2002 5 NaN NaN NaN NaN
[35549 rows x 9 columns]OUTPUT
species_id genus species taxa
0 AB Amphispiza bilineata Bird
1 AH Ammospermophilus harrisi Rodent
2 AS Ammodramus savannarum Bird
3 BA Baiomys taylori Rodent
4 CB Campylorhynchus brunneicapillus Bird
.. ... ... ... ...
49 UP Pipilo sp. Bird
50 UR Rodent sp. Rodent
51 US Sparrow sp. Bird
52 ZL Zonotrichia leucophrys Bird
53 ZM Zenaida macroura Bird
[54 rows x 4 columns]Concatenating DataFrames
We can use the concat function in pandas to append
either columns or rows from one DataFrame to another. Let’s grab two
subsets of our data to see how this works.
PYTHON
# Read in first 10 lines of surveys table
survey_sub = surveys_df.head(10)
# Grab the last 10 rows
survey_sub_last10 = surveys_df.tail(10)
# Reset the index values to the second dataframe appends properly
survey_sub_last10 = survey_sub_last10.reset_index(drop=True)
# drop=True option avoids adding new index column with old index valuesWhen we concatenate DataFrames, we need to specify the axis.
axis=0 tells pandas to stack the second DataFrame UNDER the
first one. It will automatically detect whether the column names are the
same and will stack accordingly. axis=1 will stack the
columns in the second DataFrame to the RIGHT of the first DataFrame. To
stack the data vertically, we need to make sure we have the same columns
and associated column format in both datasets. When we stack
horizontally, we want to make sure what we are doing makes sense
(i.e. the data are related in some way).
PYTHON
# Stack the DataFrames on top of each other
vertical_stack = pd.concat([survey_sub, survey_sub_last10], axis=0)
# Place the DataFrames side by side
horizontal_stack = pd.concat([survey_sub, survey_sub_last10], axis=1)Row Index Values and Concat
Have a look at the vertical_stack DataFrame. Notice
anything unusual? The row indexes for the two DataFrames
survey_sub and survey_sub_last10 have been
repeated. We can reindex the new DataFrame using the
reset_index() method.
Writing Out Data to CSV
We can use the to_csv command to export a DataFrame in
CSV format. Note that the code below will by default save the data into
the current working directory. We can save it to a different folder by
adding the foldername and a slash to the file
vertical_stack.to_csv('foldername/out.csv'). We use the
index=False so that pandas doesn’t include the index number
for each line.
Challenge - Combine Data
In the data folder, there is another folder called
yearly_files that contains survey data broken down into
individual files by year. Read the data from two of these files,
surveys2001.csv and surveys2002.csv, into
pandas and combine the files to make one new DataFrame. Export your
results as a CSV and make sure it reads back into pandas properly.
Joining DataFrames
When we concatenated our DataFrames, we simply added them to each other - stacking them either vertically or side by side. Another way to combine DataFrames is to use columns in each dataset that contain common values (a common unique identifier). Combining DataFrames using a common field is called “joining”. The columns containing the common values are called “join key(s)”. Joining DataFrames in this way is often useful when one DataFrame is a “lookup table” containing additional data that we want to include in the other.
NOTE: This process of joining tables is similar to what we do with tables in an SQL database.
For example, the species.csv file that we’ve been
working with is a lookup table. This table contains the genus, species
and taxa code for 55 species. The species code is unique for each line.
These species are identified in our survey data as well using the unique
species code. Rather than adding three more columns for the genus,
species and taxa to each of the 35,549 line survey
DataFrame, we can maintain the shorter table with the species
information. When we want to access that information, we can create a
query that joins the additional columns of information to the
survey DataFrame.
Storing data in this way has many benefits.
- It ensures consistency in the spelling of species attributes (genus, species and taxa) given each species is only entered once. Imagine the possibilities for spelling errors when entering the genus and species thousands of times!
- It also makes it easy for us to make changes to the species information once without having to find each instance of it in the larger survey data.
- It optimizes the size of our data.
Joining Two DataFrames
To better understand joins, let’s grab the first 10 lines of our data
as a subset to work with. We’ll use the .head() method to
do this. We’ll also read in a subset of the species table.
PYTHON
# Read in first 10 lines of surveys table
survey_sub = surveys_df.head(10)
# Import a small subset of the species data designed for this part of the lesson.
# It is stored in the data folder.
species_sub = pd.read_csv('../data/speciesSubset.csv')In this example, species_sub is the lookup table
containing genus, species, and taxa names that we want to join with the
data in survey_sub to produce a new DataFrame that contains
all of the columns from both species_df and
survey_df.
Identifying join keys
To identify appropriate join keys we first need to know which field(s) are shared between the files (DataFrames). We might inspect both DataFrames to identify these columns. If we are lucky, both DataFrames will have columns with the same name that also contain the same data. If we are less lucky, we need to identify a (differently-named) column in each DataFrame that contains the same information.
OUTPUT
Index(['species_id', 'genus', 'species', 'taxa'], dtype='object')OUTPUT
Index(['record_id', 'month', 'day', 'year', 'plot_id', 'species_id',
'sex', 'hindfoot_length', 'weight'], dtype='object')In our example, the join key is the column containing the two-letter
species identifier, which is called species_id.
Now that we know the fields with the common species ID attributes in each DataFrame, we are almost ready to join our data. However, since there are different types of joins, we also need to decide which type of join makes sense for our analysis.
Inner joins
The most common type of join is called an inner join. An inner join combines two DataFrames based on a join key and returns a new DataFrame that contains only those rows that have matching values in both of the original DataFrames. An example of an inner join, adapted from Jeff Atwood’s blogpost about SQL joins is below:
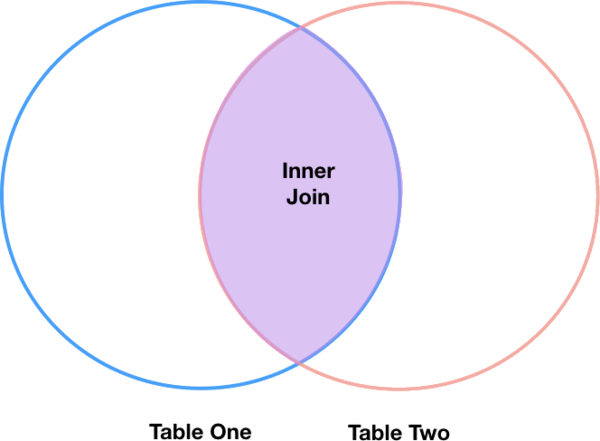
The pandas function for performing joins is called merge
and an Inner join is the default option:
PYTHON
merged_inner = pd.merge(left=survey_sub, right=species_sub, left_on='species_id', right_on='species_id')In this case, species_id is the only column name in both
DataFrames, so if we skipped the left_on and
right_on arguments, pandas would guess that we
wanted to use that column to join. However, it is usually better to be
explicit.
So what is the size of the output data?
OUTPUT
(8, 12)OUTPUT
record_id month day year plot_id species_id sex hindfoot_length \
0 1 7 16 1977 2 NL M 32
1 2 7 16 1977 3 NL M 33
2 3 7 16 1977 2 DM F 37
3 4 7 16 1977 7 DM M 36
4 5 7 16 1977 3 DM M 35
5 8 7 16 1977 1 DM M 37
6 9 7 16 1977 1 DM F 34
7 7 7 16 1977 2 PE F NaN
weight genus species taxa
0 NaN Neotoma albigula Rodent
1 NaN Neotoma albigula Rodent
2 NaN Dipodomys merriami Rodent
3 NaN Dipodomys merriami Rodent
4 NaN Dipodomys merriami Rodent
5 NaN Dipodomys merriami Rodent
6 NaN Dipodomys merriami Rodent
7 NaN Peromyscus eremicus RodentThe result of an inner join of survey_sub and
species_sub is a new DataFrame that contains the combined
set of columns from survey_sub and
species_sub. It only contains rows that have
two-letter species codes that are the same in both the
survey_sub and species_sub DataFrames. In
other words, if a row in survey_sub has a value of
species_id that does not appear in the
species_id column of species, it will not be
included in the DataFrame returned by an inner join. Similarly, if a row
in species_sub has a value of species_id that
does not appear in the species_id column of
survey_sub, that row will not be included in the DataFrame
returned by an inner join.
The two DataFrames that we want to join are passed to the
merge function using the left and
right argument. The left_on='species_id'
argument tells merge to use the species_id
column as the join key from survey_sub (the
left DataFrame). Similarly , the
right_on='species_id' argument tells merge to
use the species_id column as the join key from
species_sub (the right DataFrame). For inner
joins, the order of the left and right
arguments does not matter.
The result merged_inner DataFrame contains all of the
columns from survey_sub (record_id,
month, day, etc.) as well as all the columns
from species_sub (species_id,
genus, species, and taxa).
Notice that merged_inner has fewer rows than
survey_sub. This is an indication that there were rows in
surveys_df with value(s) for species_id that
do not exist as value(s) for species_id in
species_df.
Left joins
What if we want to add information from species_sub to
survey_sub without losing any of the information from
survey_sub? In this case, we use a different type of join
called a “left outer join”, or a “left join”.
Like an inner join, a left join uses join keys to combine two
DataFrames. Unlike an inner join, a left join will return all
of the rows from the left DataFrame, even those rows whose
join key(s) do not have values in the right DataFrame. Rows
in the left DataFrame that are missing values for the join
key(s) in the right DataFrame will simply have null (i.e.,
NaN or None) values for those columns in the resulting joined
DataFrame.
Note: a left join will still discard rows from the right
DataFrame that do not have values for the join key(s) in the
left DataFrame.
A left join is performed in pandas by calling the same
merge function used for inner join, but using the
how='left' argument:
PYTHON
merged_left = pd.merge(left=survey_sub, right=species_sub, how='left', left_on='species_id', right_on='species_id')
merged_leftOUTPUT
record_id month day year plot_id species_id sex hindfoot_length \
0 1 7 16 1977 2 NL M 32
1 2 7 16 1977 3 NL M 33
2 3 7 16 1977 2 DM F 37
3 4 7 16 1977 7 DM M 36
4 5 7 16 1977 3 DM M 35
5 6 7 16 1977 1 PF M 14
6 7 7 16 1977 2 PE F NaN
7 8 7 16 1977 1 DM M 37
8 9 7 16 1977 1 DM F 34
9 10 7 16 1977 6 PF F 20
weight genus species taxa
0 NaN Neotoma albigula Rodent
1 NaN Neotoma albigula Rodent
2 NaN Dipodomys merriami Rodent
3 NaN Dipodomys merriami Rodent
4 NaN Dipodomys merriami Rodent
5 NaN NaN NaN NaN
6 NaN Peromyscus eremicus Rodent
7 NaN Dipodomys merriami Rodent
8 NaN Dipodomys merriami Rodent
9 NaN NaN NaN NaNThe result DataFrame from a left join (merged_left)
looks very much like the result DataFrame from an inner join
(merged_inner) in terms of the columns it contains.
However, unlike merged_inner, merged_left
contains the same number of rows as the original
survey_sub DataFrame. When we inspect
merged_left, we find there are rows where the information
that should have come from species_sub (i.e.,
species_id, genus, and taxa) is
missing (they contain NaN values):
OUTPUT
record_id month day year plot_id species_id sex hindfoot_length \
5 6 7 16 1977 1 PF M 14
9 10 7 16 1977 6 PF F 20
weight genus species taxa
5 NaN NaN NaN NaN
9 NaN NaN NaN NaNThese rows are the ones where the value of species_id
from survey_sub (in this case, PF) does not
occur in species_sub.
Other join types
The pandas merge function supports other join types:
- Right (outer) join: Invoked by passing
how='right'as an argument. Similar to a left join, except all rows from therightDataFrame are kept, while rows from theleftDataFrame without matching join key(s) values are discarded. - Full (outer) join: Invoked by passing
how='outer'as an argument. This join type returns the all pairwise combinations of rows from both DataFrames; i.e., the Cartesian product and the result DataFrame will useNaNwhere data is missing in one of the dataframes. This join type is very rarely used, but can be helpful to see all the qualities of both tables, including each common and duplicate column. - Self-join: Joins a data frame with itself. Self-joins can be useful when you want to, for instance, compare records within the same dataset based on a given criteria. A fuller discussion of how and when it might be useful to do so can be found in Self-Join and Cross Join in Pandas DataFrame
Final Challenges
Challenge - Distributions
Create a new DataFrame by joining the contents of the
surveys.csv and species.csv tables. Then
calculate and plot the distribution of:
- Number of unique taxa by plot
- Number of unique taxa by sex by plot
- taxa per plot (number of species of each taxa per plot):

Suggestion: It is also possible to plot the number of individuals for each taxa in each plot (stacked bar chart):
PYTHON
merged_left.groupby(["plot_id", "taxa"]).count()["record_id"].unstack().plot(kind='bar', stacked=True)
plt.legend(loc='upper center', ncol=3, bbox_to_anchor=(0.5, 1.05)) # stop the legend from overlapping with the bar plot
- taxa by sex by plot: Providing the Nan values with the M|F values (can also already be changed to ‘x’):
PYTHON
merged_left.loc[merged_left["sex"].isna(), "sex"] = 'M|F'
ntaxa_sex_site= merged_left.groupby(["plot_id", "sex"])["taxa"].nunique().reset_index(level=1)
ntaxa_sex_site = ntaxa_sex_site.pivot_table(values="taxa", columns="sex", index=ntaxa_sex_site.index)
ntaxa_sex_site.plot(kind="bar", legend=False, stacked=True)
plt.legend(loc='upper center', ncol=3, bbox_to_anchor=(0.5, 1.08),
fontsize='small', frameon=False)
Challenge - Diversity Index
In the data folder, there is a
plots.csvfile that contains information about the type associated with each plot. Use that data to summarize the number of plots by plot type.Calculate a diversity index of your choice for control vs rodent exclosure plots. The index should consider both species abundance and number of species. You might choose to use the simple biodiversity index described here which calculates diversity as: the number of species in the plot / the total number of individuals in the plot = Biodiversity index.
PYTHON
merged_site_type = pd.merge(merged_left, plot_info, on='plot_id')
# For each plot, get the number of species for each plot
nspecies_site = merged_site_type.groupby(["plot_id"])["species"].nunique().rename("nspecies")
# For each plot, get the number of individuals
nindividuals_site = merged_site_type.groupby(["plot_id"]).count()['record_id'].rename("nindiv")
# combine the two series
diversity_index = pd.concat([nspecies_site, nindividuals_site], axis=1)
# calculate the diversity index
diversity_index['diversity'] = diversity_index['nspecies']/diversity_index['nindiv']Making a bar chart from this diversity index:
- Pandas’
mergeandconcatcan be used to combine subsets of a DataFrame, or even data from different files. -
joinfunction combines DataFrames based on index or column. - Joining two DataFrames can be done in multiple ways (left, right, and inner) depending on what data must be in the final DataFrame.
-
to_csvcan be used to write out DataFrames in CSV format.
Content from Data Workflows and Automation
Last updated on 2025-11-04 | Edit this page
Overview
Questions
- Can I automate operations in Python?
- What are functions and why should I use them?
Objectives
- Describe why for loops are used in Python.
- Employ for loops to automate data analysis.
- Write unique filenames in Python.
- Build reusable code in Python.
- Write functions using conditional statements (if, then, else).
So far, we’ve used Python and the pandas library to explore and manipulate individual datasets by hand, much like we would do in a spreadsheet. The beauty of using a programming language like Python, though, comes from the ability to automate data processing through the use of loops and functions.
For loops
Loops allow us to repeat a workflow (or series of actions) a given number of times or while some condition is true. We would use a loop to automatically process data that’s stored in multiple files (daily values with one file per year, for example). Loops lighten our work load by performing repeated tasks without our direct involvement and make it less likely that we’ll introduce errors by making mistakes while processing each file by hand.
Let’s write a simple for loop that simulates what a kid might see during a visit to the zoo:
OUTPUT
['lion', 'tiger', 'crocodile', 'vulture', 'hippo']OUTPUT
lion
tiger
crocodile
vulture
hippoThe line defining the loop must start with for and end
with a colon, and the body of the loop must be indented.
In this example, creature is the loop variable that
takes the value of the next entry in animals every time the
loop goes around. We can call the loop variable anything we like. For
example, we could change the loop variable name to x, and it’d still
work. After the loop finishes, the loop variable will still exist and
will have the value of the last entry in the collection:
PYTHON
animals = ['lion', 'tiger', 'crocodile', 'vulture', 'hippo']
for x in animals:
print('The current value of the loop variable is: ' + x)
print('At the end of the loop, the loop variable is: ' + x)OUTPUT
The current value of the loop variable is: lion
The current value of the loop variable is: tiger
The current value of the loop variable is: crocodile
The current value of the loop variable is: vulture
The current value of the loop variable is: hippo
At the end of the loop, the loop variable is: hippoWe are not asking Python to print the value of the loop variable
anymore, but the for loop still runs and the value of x
changes on each pass through the loop.
Challenge - Loops
Rewrite the previous loop so you get a counter of each iteration of the loop. The output should look similar to this:
OUTPUT
0 - The current value of the loop variable is: lion
1 - The current value of the loop variable is: tiger
2 - The current value of the loop variable is: crocodile
3 - The current value of the loop variable is: vulture
4 - The current value of the loop variable is: hippo
At the end of the loop, the loop variable is: hippoAutomating data processing using For Loops
The file we’ve been using so far, surveys.csv, contains
25 years of data and is very large. We would like to separate the data
for each year into a separate file.
Let’s start by making a new directory inside the folder
data to store all of these files using the module
os:
The command os.makedirs is equivalent to
mkdir in the shell. The exist_ok = True will
avoid a FileExistsError being raised, in case we run the code cell
multiple times. Just so we are sure, we can check that the new directory
was created within the data folder:
OUTPUT
['plots.csv',
'portal_mammals.sqlite',
'species.csv',
'survey2001.csv',
'survey2002.csv',
'surveys.csv',
'surveys2002_temp.csv',
'yearly_files']The command os.listdir is equivalent to ls
in the shell.
In previous lessons, we saw how to use the library pandas to load the species data into memory as a DataFrame, how to select a subset of the data using some criteria, and how to write the DataFrame into a CSV file. Let’s write a script that performs those three steps in sequence for the year 2002:
PYTHON
import pandas as pd
# Load the data into a DataFrame
surveys_df = pd.read_csv('../data/surveys.csv')
# Select only data for the year 2002
surveys2002 = surveys_df[surveys_df.year == 2002]
# Write the new DataFrame to a CSV file
surveys2002.to_csv('../data/yearly_files/surveys2002.csv')To create yearly data files, we could repeat the last two commands over and over, once for each year of data. Repeating code is neither elegant nor practical, and is very likely to introduce errors into your code. We want to turn what we’ve just written into a loop that repeats the last two commands for every year in the dataset.
Let’s start by writing a loop that prints the names of the files we want to create - the dataset we are using covers 1977 through 2002, and we’ll create a separate file for each of those years. Listing the filenames is a good way to confirm that the loop is behaving as we expect.
We have seen that we can loop over a list of items, so we need a list of years to loop over. We can get the years in our DataFrame with:
OUTPUT
0 1977
1 1977
2 1977
3 1977
...
35545 2002
35546 2002
35547 2002
35548 2002but we want only unique years, which we can get using the
unique method which we have already seen.
OUTPUT
array([1977, 1978, 1979, 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987,
1988, 1989, 1990, 1991, 1992, 1993, 1994, 1995, 1996, 1997, 1998,
1999, 2000, 2001, 2002], dtype=int64)Putting this into our for loop we get
PYTHON
for year in surveys_df['year'].unique():
filename='../data/yearly_files/surveys' + str(year) + '.csv'
print(filename)OUTPUT
../data/yearly_files/surveys1977.csv
../data/yearly_files/surveys1978.csv
../data/yearly_files/surveys1979.csv
../data/yearly_files/surveys1980.csv
../data/yearly_files/surveys1981.csv
../data/yearly_files/surveys1982.csv
../data/yearly_files/surveys1983.csv
../data/yearly_files/surveys1984.csv
../data/yearly_files/surveys1985.csv
../data/yearly_files/surveys1986.csv
../data/yearly_files/surveys1987.csv
../data/yearly_files/surveys1988.csv
../data/yearly_files/surveys1989.csv
../data/yearly_files/surveys1990.csv
../data/yearly_files/surveys1991.csv
../data/yearly_files/surveys1992.csv
../data/yearly_files/surveys1993.csv
../data/yearly_files/surveys1994.csv
../data/yearly_files/surveys1995.csv
../data/yearly_files/surveys1996.csv
../data/yearly_files/surveys1997.csv
../data/yearly_files/surveys1998.csv
../data/yearly_files/surveys1999.csv
../data/yearly_files/surveys2000.csv
../data/yearly_files/surveys2001.csv
../data/yearly_files/surveys2002.csvWe can now add the rest of the steps we need to create separate text files:
PYTHON
# Load the data into a DataFrame
surveys_df = pd.read_csv('../data/surveys.csv')
for year in surveys_df['year'].unique():
# Select data for the year
surveys_year = surveys_df[surveys_df.year == year]
# Write the new DataFrame to a CSV file
filename = '../data/yearly_files/surveys' + str(year) + '.csv'
surveys_year.to_csv(filename)Look inside the yearly_files directory and check a
couple of the files you just created to confirm that everything worked
as expected.
Writing Unique File Names
Notice that the code above created a unique filename for each year.
Let’s break down the parts of this name:
- The first part is some text that specifies the directory to store
our data file in (../data/yearly_files/) and the first part of the file
name (surveys):
'../data/yearly_files/surveys' - We can concatenate this with the value of a variable, in this case
yearby using the plus+sign and the variable we want to add to the file name:+ str(year) - Then we add the file extension as another text string:
+ '.csv'
Notice that we use single quotes to add text strings. The variable is
not surrounded by quotes. This code produces the string
../data/yearly_files/surveys2002.csv which contains the
path to the new filename AND the file name itself.
Challenge - Modifying loops
Some of the surveys you saved are missing data (they have null values that show up as NaN - Not A Number - in the DataFrames and do not show up in the text files). Modify the for loop so that the entries with null values are not included in the yearly files.
Let’s say you only want to look at data from a given multiple of years. How would you modify your loop in order to generate a data file for only every 5th year, starting from 1977?
Instead of splitting out the data by years, a colleague wants to do analyses each species separately. How would you write a unique CSV file for each species?
-
You could just make a list manually, however, why not check the first and last year making use of the code itself?
Building reusable and modular code with functions
Suppose that separating large data files into individual yearly files is a task that we frequently have to perform. We could write a for loop like the one above every time we needed to do it but that would be time consuming and error prone. A more elegant solution would be to create a reusable tool that performs this task with minimum input from the user. To do this, we are going to turn the code we’ve already written into a function.
Functions are reusable, self-contained pieces of code that are called with a single command. They can be designed to accept arguments as input and return values, but they don’t need to do either. Variables declared inside functions only exist while the function is running and if a variable within the function (a local variable) has the same name as a variable somewhere else in the code, the local variable hides but doesn’t overwrite the other.
Every method used in Python (for example, print) is a
function, and the libraries we import (say, pandas) are a
collection of functions. We will only use functions that are housed
within the same code that uses them, but we can also write functions
that can be used by different programs.
Functions are declared following this general structure:
PYTHON
def this_is_the_function_name(input_argument1, input_argument2):
# The body of the function is indented
# This function prints the two arguments to screen
print('The function arguments are:', input_argument1, input_argument2, '(this is done inside the function!)')
# And returns their product
return input_argument1 * input_argument2The function declaration starts with the word def,
followed by the function name and any arguments in parenthesis, and ends
in a colon. The body of the function is indented just like loops are. If
the function returns something when it is called, it includes a return
statement at the end.
This is how we call the function:
OUTPUT
The function arguments are: 2 5 (this is done inside the function!)OUTPUT
Their product is: 10 (this is done outside the function!)Challenge - Functions
- Change the values of the arguments provided to the function and check its output
- Try calling the function by giving it the wrong number of arguments
(not 2) or not assigning the function call to a variable (no
product_of_inputs =) - Declare a variable inside the function and test to see where it exists (Hint: can you print it from outside the function?)
- Explore what happens when a variable both inside and outside the function have the same name. What happens to the global variable when you change the value of the local variable?
-
For example, using an integer and a string:
OUTPUT
The function arguments are: 3 clap (this is done inside the function!) clapclapclap -
For example, providing a third integer argument:
ERROR
TypeError: this_is_the_function_name() takes 2 positional arguments but 3 were givenand proving the correct number of arguments but not capturing the return value in a variable:
OUTPUT
The function arguments are: 23 108 (this is done inside the function!) 2484 -
An example looking at variables defined outside and inside the function. Note that the explanatory comments have been removed from the function definition.
PYTHON
var_defined_outside = 'outside' def this_is_the_function_name(input_argument1, input_argument2): var_defined_inside = 'inside' print('The function arguments are:', input_argument1, input_argument2, '(this is done inside the function!)') print('This variable was created ' + var_defined_outside + ' the function') return input_argument1 * input_argument2 this_is_the_function_name(3.3, 7.9) print('This variable was created' + var_defined_inside + ' the function')OUTPUT
The function arguments are: 3.3 7.9 (this is done inside the function!) This variable was created outside the function 26.07ERROR
NameError: name 'var_defined_inside' is not definedWe can see from these results that variables defined outside the function are available for use inside them, while variables defined inside a function are not available outside them.
-
PYTHON
shared_variable_name = 'who would I be' def this_is_the_function_name(input_argument1, input_argument2): shared_variable_name = 'without you' print('The function arguments are:', input_argument1, input_argument2, '(this is done inside the function!)') return input_argument1 * input_argument2 print(shared_variable_name) this_is_the_function_name(2, 3) # does calling the function change the variable's value? print(shared_variable_name)OUTPUT
who would I be The function arguments are: 2 3 (this is done inside the function!) 6 who would I beAnd what about if we modify the variable inside the function?
PYTHON
def this_is_the_function_name(input_argument1, input_argument2): shared_variable_name = 'without you' shared_variable_name = shared_variable_name + ', without them?' print(shared_variable_name) print('The function arguments are:', input_argument1, input_argument2, '(this is done inside the function!)') return input_argument1 * input_argument2 this_is_the_function_name(2, 3) print(shared_variable_name)OUTPUT
without you, without them? The function arguments are: 2 3 (this is done inside the function!) 6 who would I be
We can now turn our code for saving yearly data files into a function. There are many different “chunks” of this code that we can turn into functions, and we can even create functions that call other functions inside them. Let’s first write a function that separates data for just one year and saves that data to a file:
PYTHON
def one_year_csv_writer(this_year, all_data):
"""
Writes a csv file for data from a given year.
this_year -- year for which data is extracted
all_data -- DataFrame with multi-year data
"""
# Select data for the year
surveys_year = all_data[all_data.year == this_year]
# Write the new DataFrame to a csv file
filename = '../data/yearly_files/function_surveys' + str(this_year) + '.csv'
surveys_year.to_csv(filename)The text between the two sets of triple double quotes is called a
docstring and contains the documentation for the function. It does
nothing when the function is running and is therefore not necessary, but
it is good practice to include docstrings as a reminder of what the code
does. Docstrings in functions also become part of their ‘official’
documentation, and we can see them by typing
help(function_name):
OUTPUT
Help on function one_year_csv_writer in module __main__:
one_year_csv_writer(this_year, all_data)
Writes a csv file for data from a given year.
this_year -- year for which data is extracted
all_data -- DataFrame with multi-year dataOr, when working in the Jupyter environment, adding a ?
(question mark) after the function name:
We changed the root of the name of the CSV file so we can distinguish
it from the one we wrote before. Check the yearly_files
directory for the file. Did it do what you expect?
What we really want to do, though, is create files for multiple years
without having to request them one by one. Let’s write another function
that replaces the entire for loop by looping through a
sequence of years and repeatedly calling the function we just wrote,
one_year_csv_writer:
PYTHON
def yearly_data_csv_writer(start_year, end_year, all_data):
"""
Writes separate CSV files for each year of data.
start_year -- the first year of data we want
end_year -- the last year of data we want
all_data -- DataFrame with multi-year data
"""
# "end_year" is the last year of data we want to pull, so we loop to end_year+1
for year in range(start_year, end_year+1):
one_year_csv_writer(year, all_data)Because people will naturally expect that the end year for the files
is the last year with data, the for loop inside the function ends at
end_year + 1. By writing the entire loop into a function,
we’ve made a reusable tool for whenever we need to break a large data
file into yearly files. Because we can specify the first and last year
for which we want files, we can even use this function to create files
for a subset of the years available. This is how we call this
function:
PYTHON
# Load the data into a DataFrame
surveys_df = pd.read_csv('../data/surveys.csv')
# Create CSV files
yearly_data_csv_writer(1977, 2002, surveys_df)BEWARE! If you are using Jupyter Notebooks and you modify a function, you MUST re-run that cell in order for the changed function to be available to the rest of the code. Nothing will visibly happen when you do this, though, because defining a function without calling it doesn’t produce an output. Any cells that use the now-changed functions will also have to be re-run for their output to change.
Challenge: More functions
- Add two arguments to the functions we wrote that take the path of the directory where the files will be written and the root of the file name. Create a new set of files with a different name in a different directory.
- How could you use the function
yearly_data_csv_writerto create a CSV file for only one year? (Hint: think about the syntax forrange) - Make the functions return a list of the files they have written.
There are many ways you can do this (and you should try them all!):
either of the functions can print to screen, either can use a return
statement to give back numbers or strings to their function call, or you
can use some combination of the two. You could also try using the
oslibrary to list the contents of directories. - Explore what happens when variables are declared inside each of the functions versus in the main (non-indented) body of your code. What is the scope of the variables (where are they visible)? What happens when they have the same name but are given different values?
PYTHON
def one_year_csv_writer(this_year, all_data, folder_to_save, root_name): """ Writes a csv file for data from a given year. Parameters --------- this_year : int year for which data is extracted all_data: pd.DataFrame DataFrame with multi-year data folder_to_save : str folder to save the data files root_name: str root of the filenames to save the data """ # Select data for the year surveys_year = all_data[all_data.year == this_year] # Write the new DataFrame to a csv file filename = os.path.join(folder_to_save, ''.join([root_name, str(this_year), '.csv'])) surveys_year.to_csv(filename) start_year = 1978 end_year = 2000 directory_name = 'different_directory' root_file_name = 'different_file_name' for year in range(start_year, end_year+1): one_year_csv_writer(year, surveys_df, directory_name, root_file_name)yearly_data_csv_writer(2000, 2000, surveys_df)-
Building a list and returning it:
PYTHON
def one_year_csv_writer(this_year, all_data): """ Writes a csv file for data from a given year. this_year -- year for which data is extracted all_data -- DataFrame with multi-year data """ # Select data for the year surveys_year = all_data[all_data.year == this_year] # Write the new DataFrame to a csv file filename = '../data/yearly_files/function_surveys' + str(this_year) + '.csv' surveys_year.to_csv(filename) return filename def yearly_data_csv_writer(start_year, end_year, all_data): """ Writes separate CSV files for each year of data. start_year -- the first year of data we want end_year -- the last year of data we want all_data -- DataFrame with multi-year data """ # "end_year" is the last year of data we want to pull, so we loop to end_year+1 output_files = [] for year in range(start_year, end_year+1): output_files.append(one_year_csv_writer(year, all_data)) return output_files print(yearly_data_csv_writer(2000, 2001, surveys_df))OUTPUT
['../data/yearly_files/function_surveys2000.csv', '../data/yearly_files/function_surveys2001.csv']
The functions we wrote demand that we give them a value for every
argument. Ideally, we would like these functions to be as flexible and
independent as possible. Let’s modify the function
yearly_data_csv_writer so that the start_year
and end_year default to the full range of the data if they
are not supplied by the user. Arguments can be given default values with
an equal sign in the function declaration. Any arguments in the function
without default values (here, all_data) is a required
argument and MUST come before the argument with default values (which
are optional in the function call).
PYTHON
def yearly_data_arg_test(all_data, start_year=1977, end_year=2002):
"""
Modified from yearly_data_csv_writer to test default argument values!
start_year -- the first year of data we want (default 1977)
end_year -- the last year of data we want (default 2002)
all_data -- DataFrame with multi-year data
"""
return start_year, end_year
start, end = yearly_data_arg_test(surveys_df, 1988, 1993)
print('Both optional arguments:\t', start, end)
start, end = yearly_data_arg_test(surveys_df)
print('Default values:\t\t\t', start, end)OUTPUT
Both optional arguments: 1988 1993
Default values: 1977 2002The “\t” in the print statements are tabs, used to make
the text align and be easier to read.
But what if our dataset doesn’t start in 1977 and end in 2002? We can modify the function so that it looks for the start and end years in the dataset if those dates are not provided:
PYTHON
def yearly_data_arg_test(all_data, start_year=None, end_year=None):
"""
Modified from yearly_data_csv_writer to test default argument values!
all_data -- DataFrame with multi-year data
start_year -- the first year of data we want, Check all_data! (default None)
end_year -- the last year of data we want; Check all_data! (default None)
"""
if start_year is None:
start_year = min(all_data.year)
if end_year is None:
end_year = max(all_data.year)
return start_year, end_year
start, end = yearly_data_arg_test(surveys_df, 1988, 1993)
print('Both optional arguments:\t', start, end)
start, end = yearly_data_arg_test(surveys_df)
print('Default values:\t\t\t', start, end)OUTPUT
Both optional arguments: 1988 1993
Default values: 1977 2002The default values of the start_year and
end_year arguments in the function
yearly_data_arg_test are now None. This is a
built-in constant in Python that indicates the absence of a value -
essentially, that the variable exists in the namespace of the function
(the directory of variable names) but that it doesn’t correspond to any
existing object.
Challenge - Variables
- What type of object corresponds to a variable declared as
None? (Hint: create a variable set toNoneand use the functiontype()) - Compare the behavior of the function
yearly_data_arg_testwhen the arguments haveNoneas a default and when they do not have default values. - What happens if you only include a value for
start_yearin the function call? Can you write the function call with only a value forend_year? (Hint: think about how the function must be assigning values to each of the arguments - this is related to the need to put the arguments without default values before those with default values in the function definition!)
-
OUTPUT
NoneType -
Removing
Noneas the default value for thestart_yearandend_yearparameters results in aTypeErrorbecause the function now expects three arguments to be provided:PYTHON
def yearly_data_arg_test(all_data, start_year, end_year): """ Modified from yearly_data_csv_writer to test default argument values! all_data -- DataFrame with multi-year data start_year -- the first year of data we want, Check all_data! (default None) end_year -- the last year of data we want; Check all_data! (default None) """ if start_year is None: start_year = min(all_data.year) if end_year is None: end_year = max(all_data.year) return start_year, end_year start, end = yearly_data_arg_test(surveys_df)ERROR
TypeError: yearly_data_arg_test() missing 2 required positional arguments: 'start_year' and 'end_year' -
Returning to the original definition of the function (i.e. with the default
Nonevalues):OUTPUT
(1985, 2002)If we want to provide only the end year, we have to name the arguments:
OUTPUT
(1977, 1985)
If Statements
The body of the test function now has two conditionals (if
statements) that check the values of start_year and
end_year. If statements execute a segment of code when some
condition is met. They commonly look something like this:
PYTHON
a = 5
if a<0: # Meets first condition?
# if a IS less than zero
print('a is a negative number')
elif a>0: # Did not meet first condition. meets second condition?
# if a ISN'T less than zero and IS more than zero
print('a is a positive number')
else: # Met neither condition
# if a ISN'T less than zero and ISN'T more than zero
print('a must be zero!')Which would return:
OUTPUT
a is a positive numberChange the value of a to see how this function works.
The statement elif means “else if”, and all of the
conditional statements must end in a colon.
The if statements in the function yearly_data_arg_test
check whether there is an object associated with the variable names
start_year and end_year. If those variables
are None, the if statements return the boolean
True and execute whatever is in their body. On the other
hand, if the variable names are associated with some value (they got a
number in the function call), the if statements return
False and do not execute. The opposite conditional
statements, which would return True if the variables were
associated with objects (if they had received value in the function
call), would be if start_year and
if end_year.
As we’ve written it so far, the function
yearly_data_arg_test associates values in the function call
with arguments in the function definition just based on their order. If
the function gets only two values in the function call, the first one
will be associated with all_data and the second with
start_year, regardless of what we intended them to be. We
can get around this problem by calling the function using keyword
arguments, where each of the arguments in the function definition is
associated with a keyword and the function call passes values to the
function using these keywords:
PYTHON
start, end = yearly_data_arg_test(surveys_df)
print('Default values:\t\t\t', start, end)
start, end = yearly_data_arg_test(surveys_df, 1988, 1993)
print('No keywords:\t\t\t', start, end)
start, end = yearly_data_arg_test(surveys_df, start_year=1988, end_year=1993)
print('Both keywords, in order:\t', start, end)
start, end = yearly_data_arg_test(surveys_df, end_year=1993, start_year=1988)
print('Both keywords, flipped:\t\t', start, end)
start, end = yearly_data_arg_test(surveys_df, start_year=1988)
print('One keyword, default end:\t', start, end)
start, end = yearly_data_arg_test(surveys_df, end_year=1993)
print('One keyword, default start:\t', start, end)OUTPUT
Default values: 1977 2002
No keywords: 1988 1993
Both keywords, in order: 1988 1993
Both keywords, flipped: 1988 1993
One keyword, default end: 1988 2002
One keyword, default start: 1977 1993Challenge - Modifying functions
Rewrite the
one_year_csv_writerandyearly_data_csv_writerfunctions to have keyword arguments with default valuesModify the functions so that they don’t create yearly files if there is no data for a given year and display an alert to the user (Hint: use conditional statements to do this. For an extra challenge, use
trystatements!)-
The code below checks to see whether a directory exists and creates one if it doesn’t. Add some code to your function that writes out the CSV files, to check for a directory to write to.
The code that you have written so far to loop through the years is good, however it is not necessarily reproducible with different datasets. For instance, what happens to the code if we have additional years of data in our CSV files? Using the tools that you learned in the previous activities, make a list of all years represented in the data. Then create a loop to process your data, that begins at the earliest year and ends at the latest year using that list.
HINT: you can create a loop with a list as follows:
for years in year_list:
PYTHON
def one_year_csv_writer(this_year, all_data, folder_to_save='./', root_name='survey'): """ Writes a csv file for data from a given year. Parameters --------- this_year : int year for which data is extracted all_data: pd.DataFrame DataFrame with multi-year data folder_to_save : str folder to save the data files root_name: str root of the filenames to save the data """ # Select data for the year surveys_year = all_data[all_data.year == this_year] # Write the new DataFrame to a csv file filename = os.path.join(folder_to_save, ''.join([root_name, str(this_year), '.csv'])) surveys_year.to_csv(filename) def yearly_data_csv_writer(all_data, start_year=None, end_year=None): """ Writes separate CSV files for each year of data. start_year -- the first year of data we want end_year -- the last year of data we want all_data -- DataFrame with multi-year data """ if start_year is None: start_year = min(all_data.year) if end_year is None: end_year = max(all_data.year) # "end_year" is the last year of data we want to pull, so we loop to end_year+1 for year in range(start_year, end_year+1): one_year_csv_writer(year, all_data)PYTHON
def one_year_csv_writer(this_year, all_data, folder_to_save='./', root_name='survey'): """ Writes a csv file for data from a given year. Parameters --------- this_year : int year for which data is extracted all_data: pd.DataFrame DataFrame with multi-year data folder_to_save : str folder to save the data files root_name: str root of the filenames to save the data """ # Select data for the year surveys_year = all_data[all_data.year == this_year] if len(surveys_year) == 0: # 'if not len(surveys_year):' will also work print('no data available for ' + this_year + ', output file not created') else: # Write the new DataFrame to a csv file filename = os.path.join(folder_to_save, ''.join([root_name, str(this_year), '.csv'])) surveys_year.to_csv(filename)-
The solution below uses
notto invert the check for the directory, so a message is printed one time at most i.e. when the directory is created:PYTHON
def one_year_csv_writer(this_year, all_data, folder_to_save='./', root_name='survey'): """ Writes a csv file for data from a given year. Parameters --------- this_year : int year for which data is extracted all_data: pd.DataFrame DataFrame with multi-year data folder_to_save : str folder to save the data files root_name: str root of the filenames to save the data """ # Select data for the year surveys_year = all_data[all_data.year == this_year] if len(surveys_year) == 0: print('no data available for ' + this_year + ', output file not created') else: if folder_to_save not in os.listdir('.'): os.mkdir(folder_to_save) print('Processed directory created') # Write the new DataFrame to a csv file filename = os.path.join(folder_to_save, ''.join([root_name, str(this_year), '.csv'])) surveys_year.to_csv(filename)The equivalent, without using
notto invert the conditional, would be: PYTHON
def yearly_data_csv_writer(all_data, yearcolumn="year", folder_to_save='./', root_name='survey'): """ Writes separate csv files for each year of data. all_data --- DataFrame with multi-year data yearcolumn --- column name containing the year of the data folder_to_save --- folder name to store files root_name --- start of the file names stored """ years = all_data[yearcolumn].unique() filenames = [] for year in years: filenames.append(one_year_csv_writer(year, all_data, folder_to_save, root_name)) return filenames
- Loops help automate repetitive tasks over sets of items.
- Loops combined with functions provide a way to process data more efficiently than we could by hand.
- Conditional statements enable execution of different operations on different data.
- Functions enable code reuse.